Modelled Stucture
Method:Homology modeling
Teplate PDB:5U70_A
Identity:86.095%
Minimized Score:-2538.384
Detail: Structure Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0532 Transporter Info | ||||
| Gene Name | KCNT1 | ||||
| Protein Name | Potassium channel subfamily T member 1 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Modelled Stucture Method:Homology modeling Teplate PDB:5U70_A Identity:86.095% Minimized Score:-2538.384 Detail: Structure Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Mus musculus (Mouse) | |||||
| Gene Name | Kcnt1 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5U70_A Sequence Length:1224 Identity:84.194% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -2566.085 kcal/mol | |||
| Ramachandra Favored | Medium |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
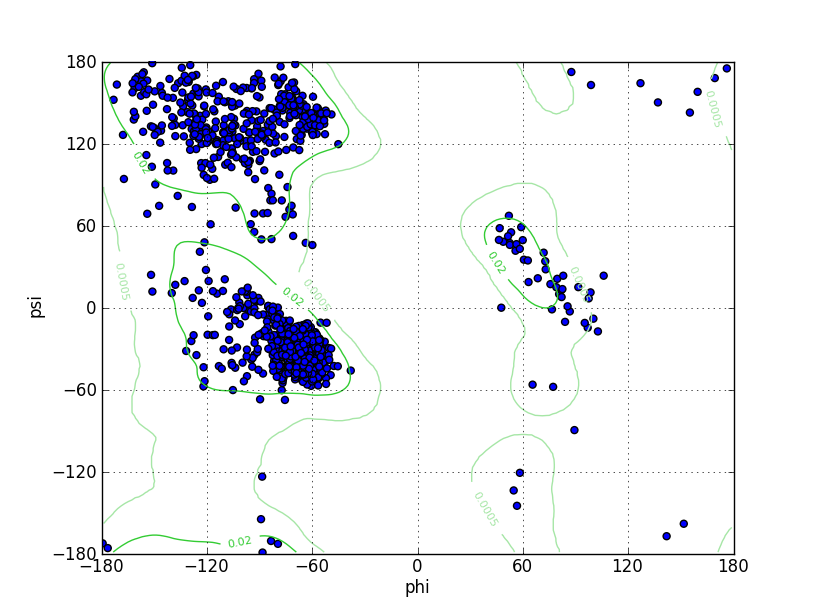
Ramz Z Score:-0.42 ±0.23 Residues in Favored Region:1186 Ramachandran favored:97.05% Number of Outliers:5 Ramachandran outliers:0.41% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
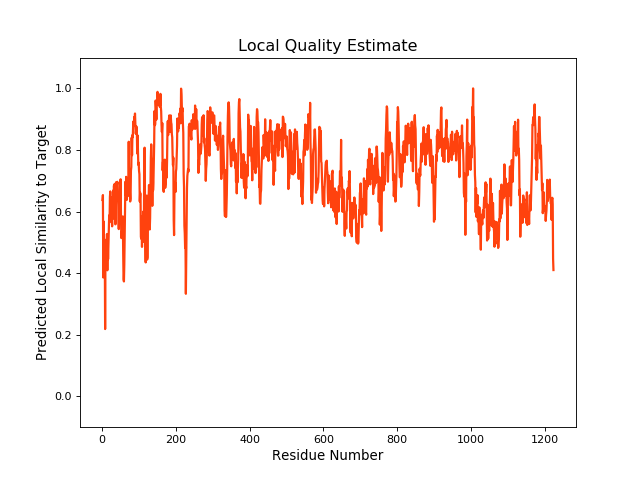
QMEANBrane Score:0.73 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Rattus norvegicus (Rat) | |||||
| Gene Name | Kcnt1 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5U70_A Sequence Length:1237 Identity:84.839% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -2597.201 kcal/mol | |||
| Ramachandra Favored | Medium |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
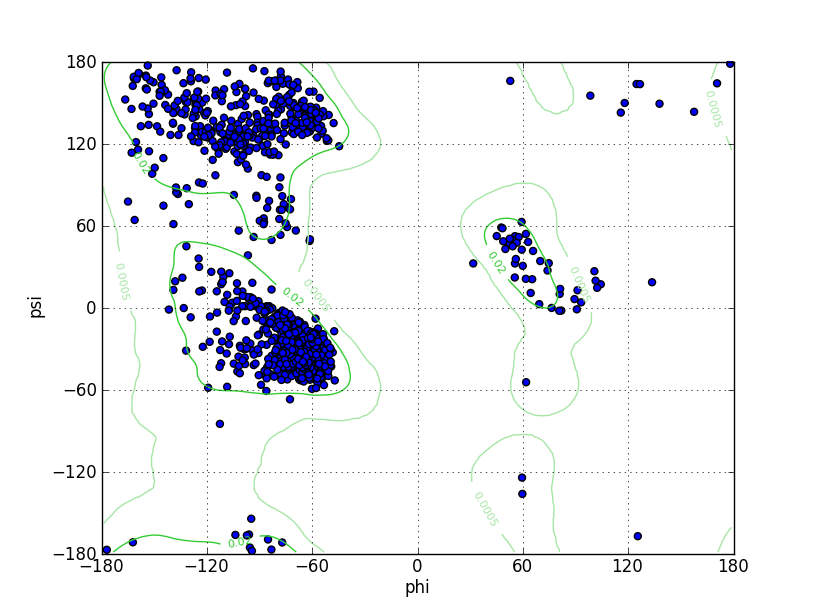
Ramz Z Score:-0.72 ±0.22 Residues in Favored Region:1199 Ramachandran favored:97.09% Number of Outliers:5 Ramachandran outliers:0.40% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
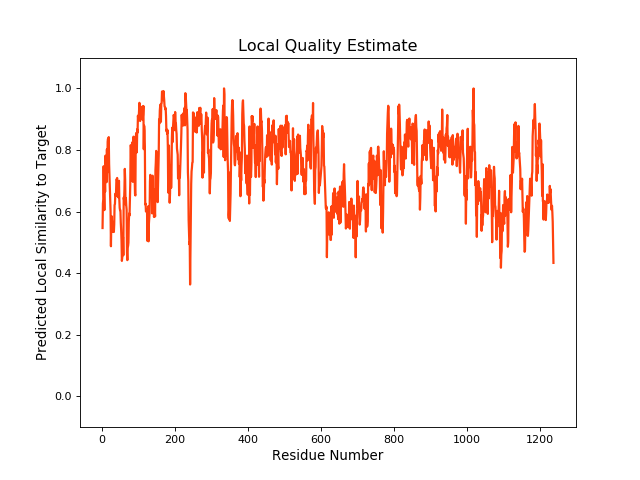
QMEANBrane Score:0.73 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Gallus gallus (Chicken) | |||||
| Gene Name | KCNT1 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure | |||||
| Click to Save PDB File in PDB Format | |||||
| Experimental Structures of This Model Oganism | |||||
| PDB ID | Scanning Method | Resolution | Expression System | Details | Ref |
| 5U76 | EM | 4.2 Å | Spodoptera frugiperda | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 1-1201 | Mutation | No | ||
| 5A6E | EM | 4.5 Å | Spodoptera frugiperda | [ 2] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | B/C | ||||
| Sequence Length | 244-337; 351-1019; 1141-1171 | Mutation | No | ||
| 5A6F | EM | 4.2 Å | Spodoptera frugiperda | [ 2] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | C | ||||
| Sequence Length | 351-1019; 1141-1171 | Mutation | No | ||
| 5A6G | EM | 4.5 Å | Spodoptera frugiperda | [ 2] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | B | ||||
| Sequence Length | 244-337 | Mutation | No | ||
| References | |||||
| 1 | Structural Titration of Slo2.2, a Na(+)-Dependent K(+) Channel. Cell. 2017 Jan 26;168(3):390-399.e11. | ||||
| 2 | Cryo-electron microscopy structure of the Slo2.2 Na(+)-activated K(+) channel. Nature. 2015 Nov 12;527(7577):198-203. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.