Modelled Stucture
Method:Homology modeling
Teplate PDB:5KK2_E
Identity:73.994%
Minimized Score:-470.632
Detail: Structure Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0524 Transporter Info | ||||
| Gene Name | CACNG3 | ||||
| Protein Name | Voltage-dependent calcium channel gamma-3 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Modelled Stucture Method:Homology modeling Teplate PDB:5KK2_E Identity:73.994% Minimized Score:-470.632 Detail: Structure Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Mus musculus (Mouse) | |||||
| Gene Name | Cacng3 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5KK2_E Sequence Length:315 Identity:73.684% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -531.115 kcal/mol | |||
| Ramachandra Favored | Medium |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |

Ramz Z Score:-0.35 ±0.47 Residues in Favored Region:305 Ramachandran favored:97.44% Number of Outliers:2 Ramachandran outliers:0.64% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
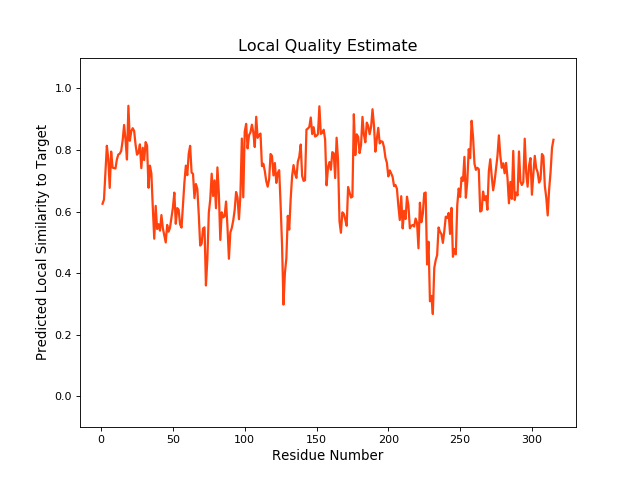
QMEANBrane Score:0.69 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Rattus norvegicus (Rat) | |||||
| Gene Name | Cacng3 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5KK2_E Sequence Length:315 Identity:73.684% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -501.791 kcal/mol | |||
| Ramachandra Favored | Medium |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
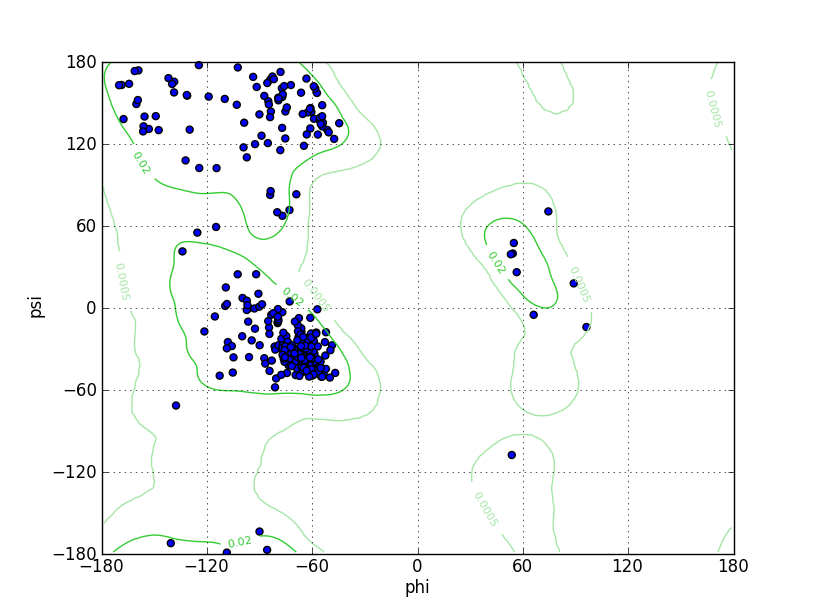
Ramz Z Score:-1.26 ±0.45 Residues in Favored Region:301 Ramachandran favored:96.17% Number of Outliers:0 Ramachandran outliers:0.00% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
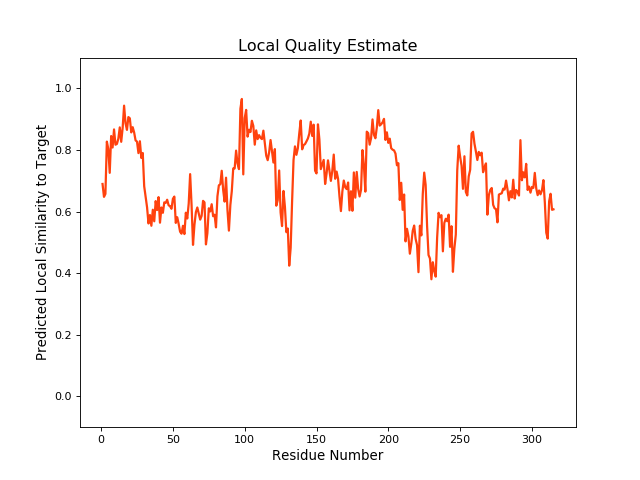
QMEANBrane Score:0.69 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Macaca fascicularis (Cynomolgus monkey) | |||||
| Gene Name | CACNG3 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5KK2_E Sequence Length:315 Identity:73.994% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -525.15 kcal/mol | |||
| Ramachandra Favored | Medium |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
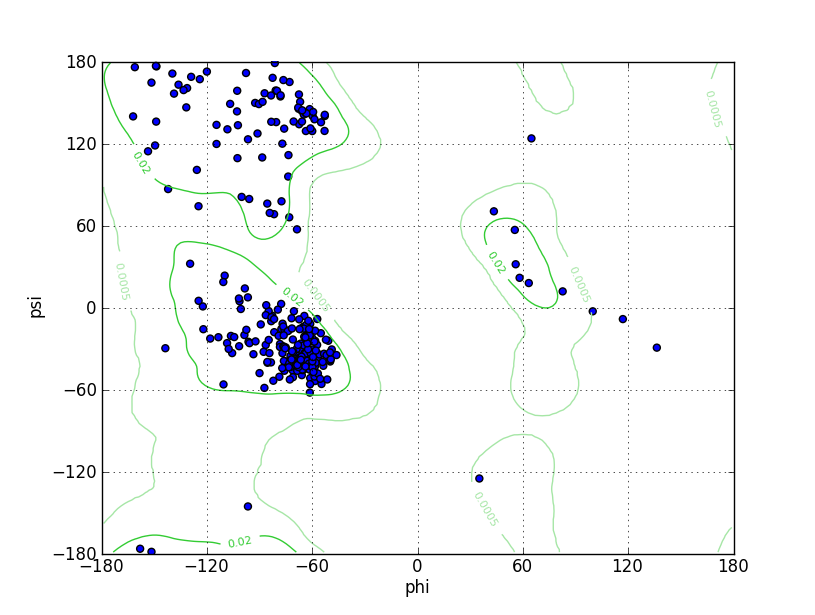
Ramz Z Score:-1.14 ±0.44 Residues in Favored Region:302 Ramachandran favored:96.49% Number of Outliers:2 Ramachandran outliers:0.64% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
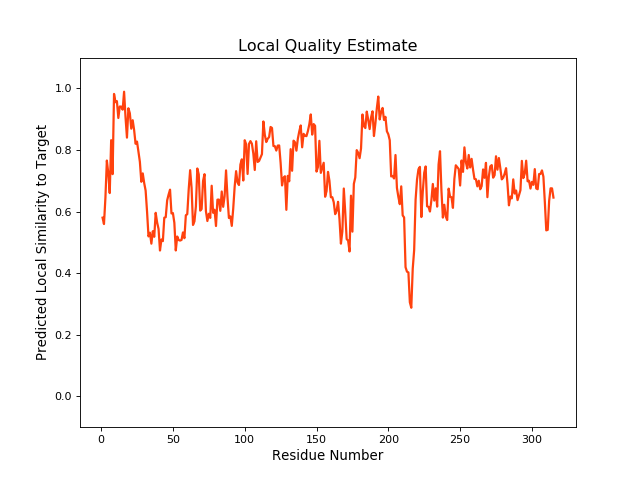
QMEANBrane Score:0.69 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| References | |||||
| 1 | Opening of glutamate receptor channel to subconductance levels. Nature. 2022;605(7908):172-178. | ||||
| 2 | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor. Nature. 2023;621(7980):877-882. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.