Modelled Stucture
Method:Homology modeling
Teplate PDB:6JPA_A
Identity:48.82%
Minimized Score:-1476.087
Detail: Structure Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0520 Transporter Info | ||||
| Gene Name | CACNA1E | ||||
| Protein Name | Voltage-gated calcium channel alpha Cav2.3 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Modelled Stucture Method:Homology modeling Teplate PDB:6JPA_A Identity:48.82% Minimized Score:-1476.087 Detail: Structure Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Mus musculus (Mouse) | |||||
| Gene Name | Cacna1e | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5GJV_A Sequence Length:846 Identity:45.195% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -1347.635 kcal/mol | |||
| Ramachandra Favored | Poor |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
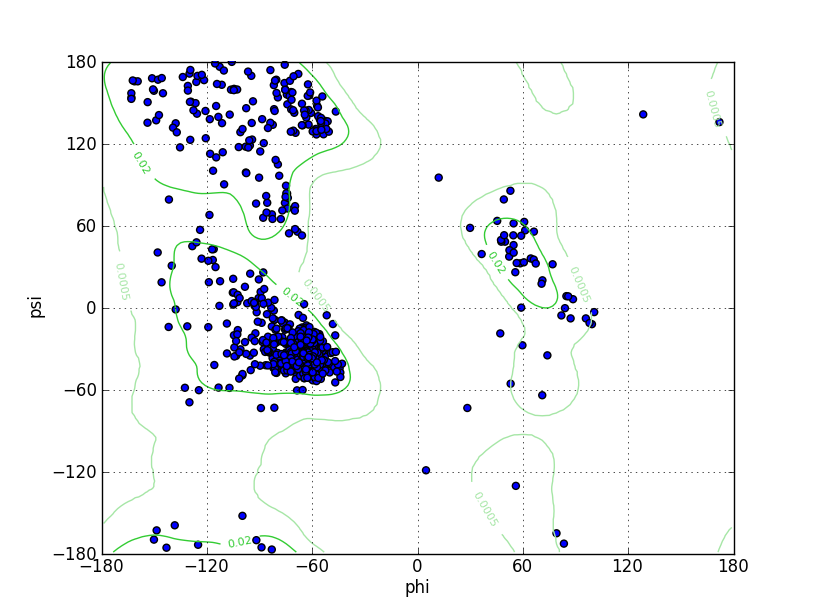
Ramz Z Score:-2.03 ±0.27 Residues in Favored Region:789 Ramachandran favored:93.48% Number of Outliers:10 Ramachandran outliers:1.18% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
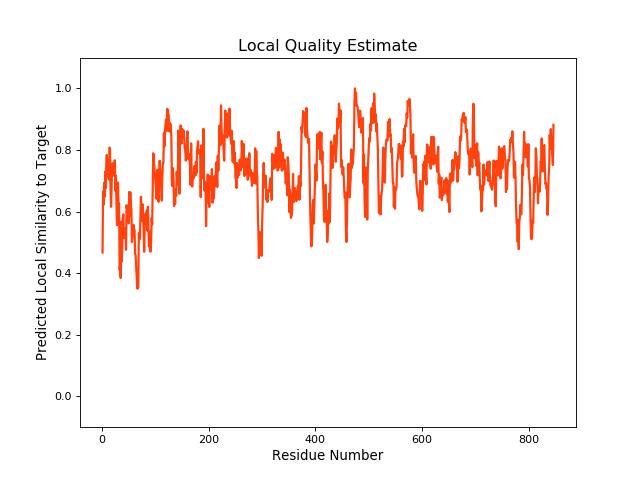
QMEANBrane Score:0.73 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Rattus norvegicus (Rat) | |||||
| Gene Name | Cacna1e | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5GJV_A Sequence Length:868 Identity:44.593% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -1236.48 kcal/mol | |||
| Ramachandra Favored | Poor |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
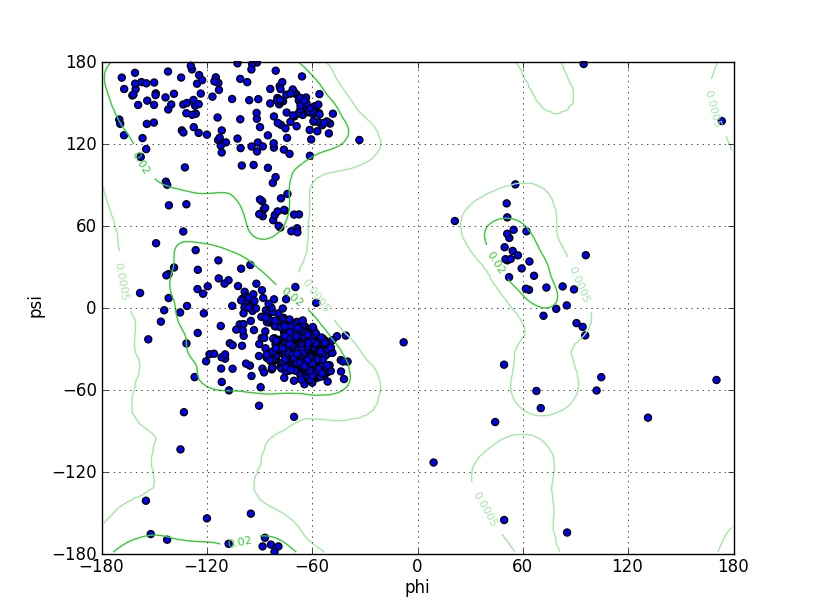
Ramz Z Score:-1.52 ±0.27 Residues in Favored Region:809 Ramachandran favored:93.42% Number of Outliers:12 Ramachandran outliers:1.39% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
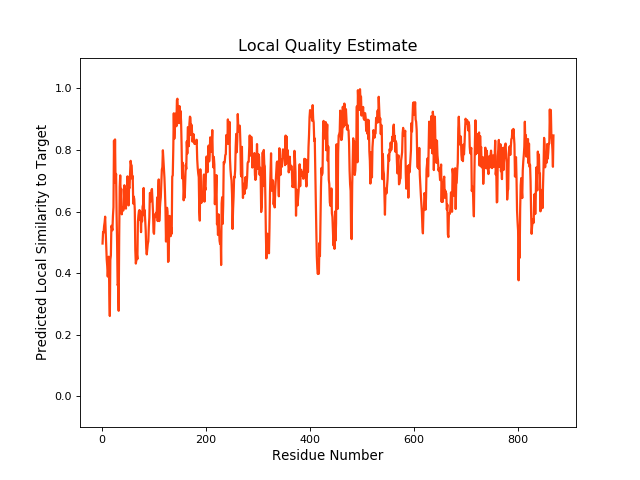
QMEANBrane Score:0.73 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Experimental Structures of This Model Oganism | |||||
| PDB ID | Scanning Method | Resolution | Expression System | Details | Ref |
| 3DVK | X-ray | 2.3 Å | Escherichia coli | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | B | ||||
| Sequence Length | 1818-1837 | Mutation | No | ||
| Oryctolagus cuniculus (Rabbit) | |||||
| Gene Name | CACNA1E | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5GJV_A Sequence Length:761 Identity:48.087% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -1431.605 kcal/mol | |||
| Ramachandra Favored | Poor |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
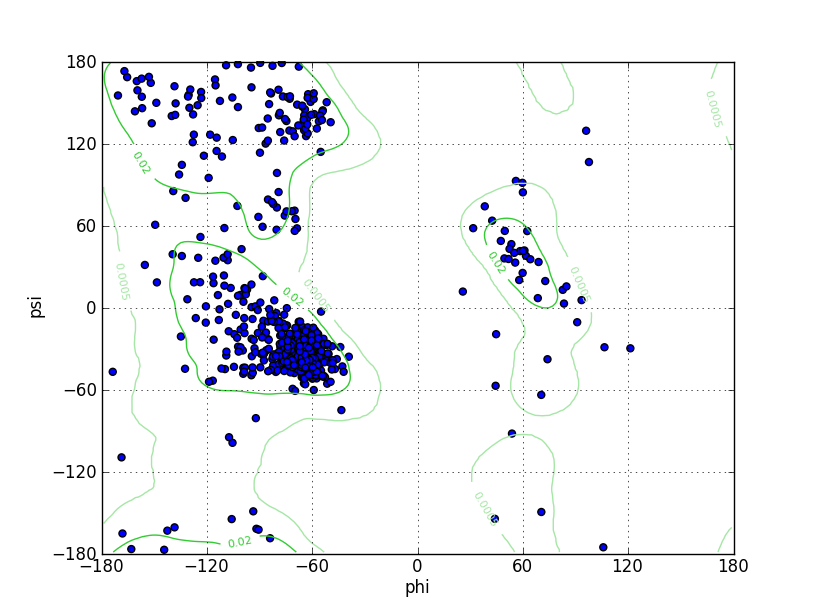
Ramz Z Score:-2.05 ±0.28 Residues in Favored Region:711 Ramachandran favored:93.68% Number of Outliers:14 Ramachandran outliers:1.84% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
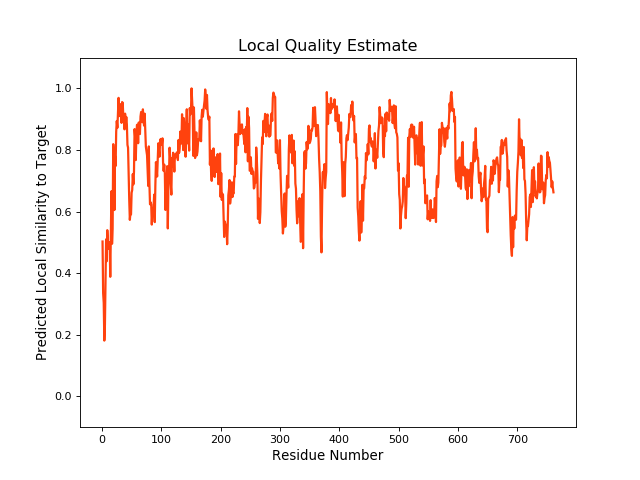
QMEANBrane Score:0.73 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| References | |||||
| 1 | Structures of CaV2 Ca2+/CaM-IQ domain complexes reveal binding modes that underlie calcium-dependent inactivation and facilitation. Structure. 2008 Oct 8;16(10):1455-67. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.