Crystal Structure of This DT
Crystallisation Method:EM
Corresponding Chain:A/B/C/D
Sequence Length:1-390
Detail: Struture Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0511 Transporter Info | ||||
| Gene Name | KCNJ11 | ||||
| Protein Name | ATP-sensitive inward rectifier potassium channel 11 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Crystal Structure of This DT Crystallisation Method:EM Corresponding Chain:A/B/C/D Sequence Length:1-390 Detail: Struture Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Cavia porcellus (Guinea pig) | |||||
| Gene Name | KCNJ11 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:6C3O_A Sequence Length:390 Identity:96.41% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -829.941 kcal/mol | |||
| Ramachandra Favored | Excellent |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
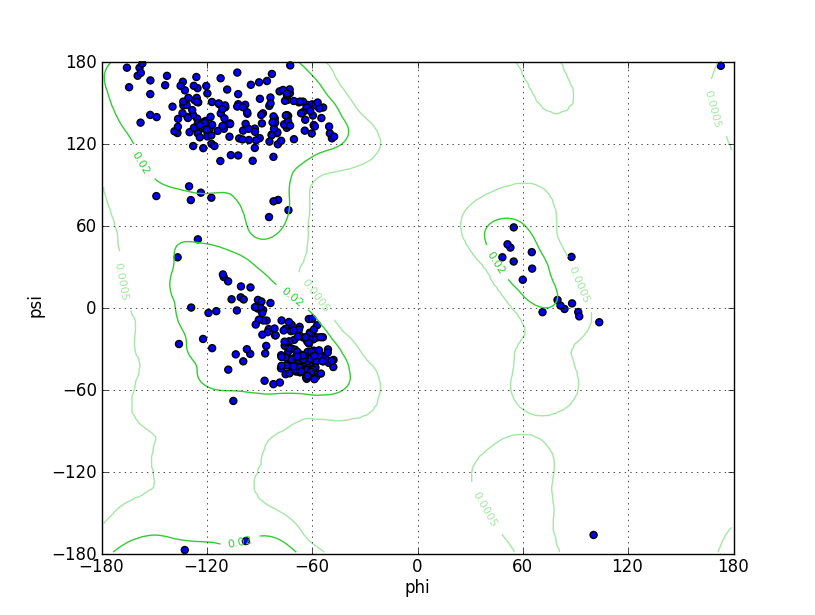
Ramz Z Score:0.05 ±0.4 Residues in Favored Region:382 Ramachandran favored:98.45% Number of Outliers:0 Ramachandran outliers:0.00% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |

QMEANBrane Score:0.75 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| References | |||||
| 1 | Ligand binding and conformational changes of SUR1 subunit in pancreatic ATP-sensitive potassium channels. Protein Cell. 2018 Jun;9(6):553-567. | ||||
| 2 | Structure of a Pancreatic ATP-Sensitive Potassium Channel. Cell. 2017 Jan 12;168(1-2):101-110.e10. | ||||
| 3 | The Structural Basis for the Binding of Repaglinide to the Pancreatic K(ATP) Channel. Cell Rep. 2019 May 7;27(6):1848-1857.e4. | ||||
| 4 | Structural insights into the mechanism of pancreatic K(ATP) channel regulation by nucleotides. Nat Commun. 2022;13(1):2770. | ||||
| 5 | Mechanism of pharmacochaperoning in a mammalian K(ATP) channel revealed by cryo-EM. Elife. 2019 Jul 25;8:e46417. | ||||
| 6 | Cryo-EM structure of the ATP-sensitive potassium channel illuminates mechanisms of assembly and gating. Elife. 2017 Jan 16;6:e24149. | ||||
| 7 | Anti-diabetic drug binding site in a mammalian K(ATP) channel revealed by Cryo-EM. Elife. 2017 Oct 24;6:e31054. | ||||
| 8 | Ligand-mediated Structural Dynamics of a Mammalian Pancreatic K(ATP) Channel. J Mol Biol. 2022;434(19):167789. | ||||
| 9 | Structure of an open K(ATP) channel reveals tandem PIP(2) binding sites mediating the Kir6.2 and SUR1 regulatory interface. Nat Commun. 2024;15(1):2502. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.