Crystal Structure of This DT
Crystallisation Method:EM
Corresponding Chain:A
Sequence Length:1-1480
Detail: Struture Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0060 Transporter Info | ||||
| Gene Name | ABCC7 | ||||
| Protein Name | Cystic fibrosis transmembrane conductance regulator | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Crystal Structure of This DT Crystallisation Method:EM Corresponding Chain:A Sequence Length:1-1480 Detail: Struture Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Equus caballus (Horse) | |||||
| Gene Name | CFTR | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5UAK_A Sequence Length:1481 Identity:94.062% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -3278.885 kcal/mol | |||
| Ramachandra Favored | Medium |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
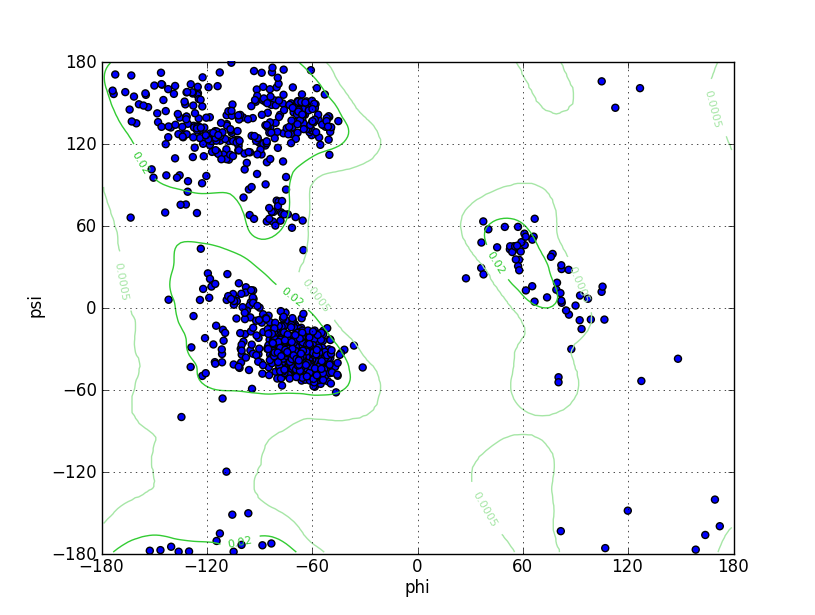
Ramz Z Score:-1.05 ±0.2 Residues in Favored Region:1444 Ramachandran favored:97.63% Number of Outliers:8 Ramachandran outliers:0.54% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
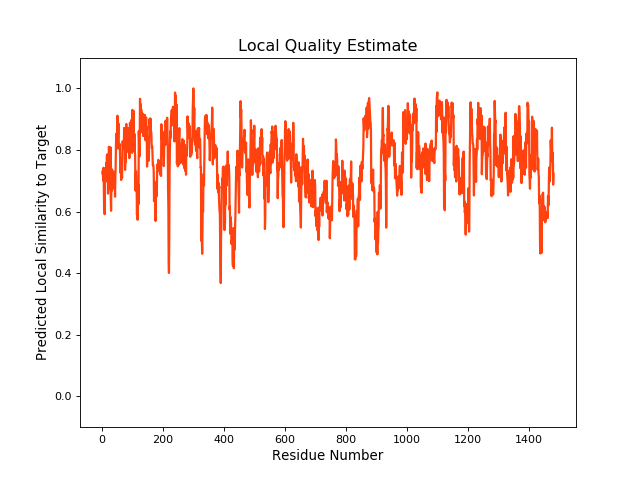
QMEANBrane Score:0.75 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Macaca mulatta (Rhesus macaque) | |||||
| Gene Name | CFTR | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5UAK_A Sequence Length:1481 Identity:98.244% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -3224.967 kcal/mol | |||
| Ramachandra Favored | Medium |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
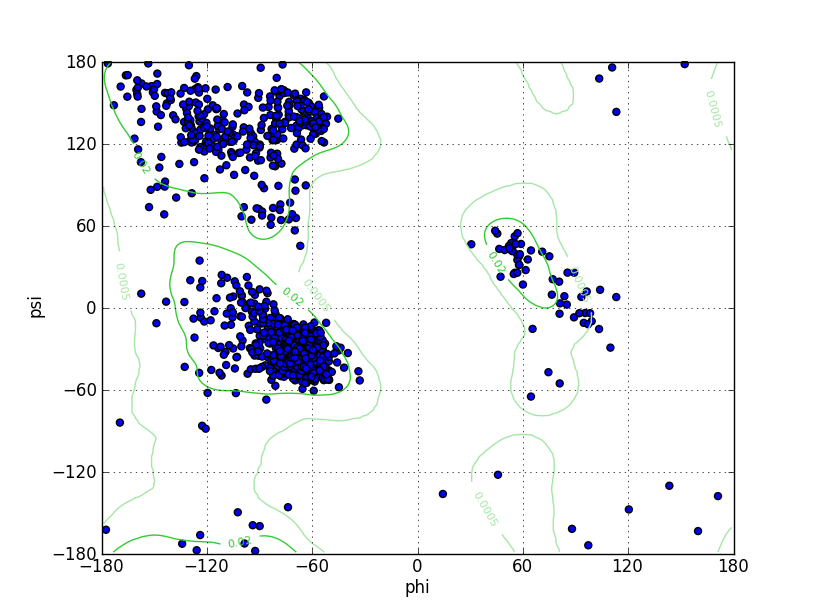
Ramz Z Score:-0.76 ±0.21 Residues in Favored Region:1435 Ramachandran favored:97.03% Number of Outliers:7 Ramachandran outliers:0.47% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |

QMEANBrane Score:0.75 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Ovis aries (Sheep) | |||||
| Gene Name | CFTR | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5UAK_A Sequence Length:1481 Identity:90.688% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -3213.509 kcal/mol | |||
| Ramachandra Favored | Medium |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
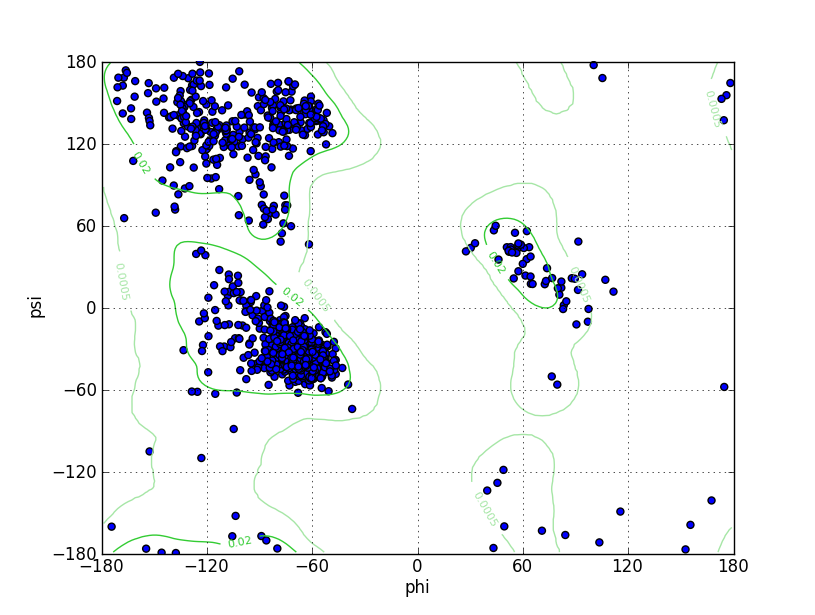
Ramz Z Score:-0.84 ±0.2 Residues in Favored Region:1437 Ramachandran favored:97.16% Number of Outliers:8 Ramachandran outliers:0.54% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
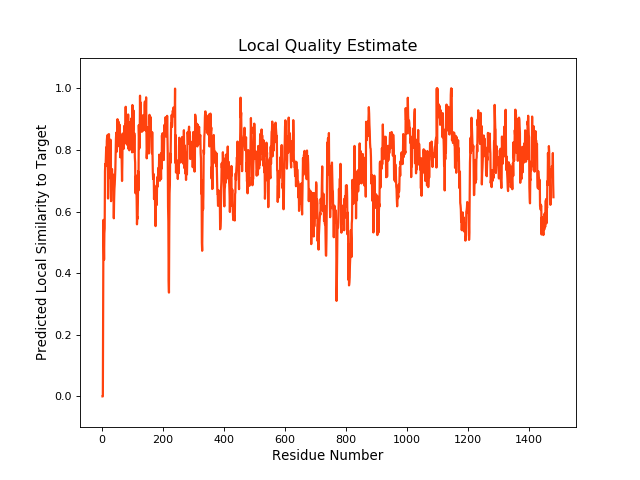
QMEANBrane Score:0.75 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Experimental Structures of This Model Oganism | |||||
| PDB ID | Scanning Method | Resolution | Expression System | Details | Ref |
| References | |||||
| 1 | Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 2004 Jan 28;23(2):282-93. | ||||
| 2 | Side chain and backbone contributions of Phe508 to CFTR folding. Nat Struct Mol Biol. 2005 Jan;12(1):10-6. | ||||
| 3 | Requirements for efficient correction of F508 CFTR revealed by analyses of evolved sequences. Cell. 2012 Jan 20;148(1-2):164-74. | ||||
| 4 | Conformational Changes of CFTR upon Phosphorylation and ATP Binding. Cell. 2017 Jul 27;170(3):483-491.e8. | ||||
| 5 | Cystic fibrosis transmembrane conductance regulator: solution structures of peptides based on the Phe508 region, the most common site of disease-causing DeltaF508 mutation. Biochemistry. 1999 Jun 8;38(23):7453-61. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.