Modelled Stucture
Method:Homology modeling
Teplate PDB:4XPB_A
Identity:55.354%
Minimized Score:-1503.772
Detail: Structure Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0455 Transporter Info | ||||
| Gene Name | SLC6A3 | ||||
| Protein Name | Sodium-dependent dopamine transporter | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Modelled Stucture Method:Homology modeling Teplate PDB:4XPB_A Identity:55.354% Minimized Score:-1503.772 Detail: Structure Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Mus musculus (Mouse) | |||||
| Gene Name | Slc6a3 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:4XPB_A Sequence Length:548 Identity:56.284% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -1531.87 kcal/mol | |||
| Ramachandra Favored | Excellent |
||||
| QMEANBrane Quality | High |
||||
| Ramachandran Plot |
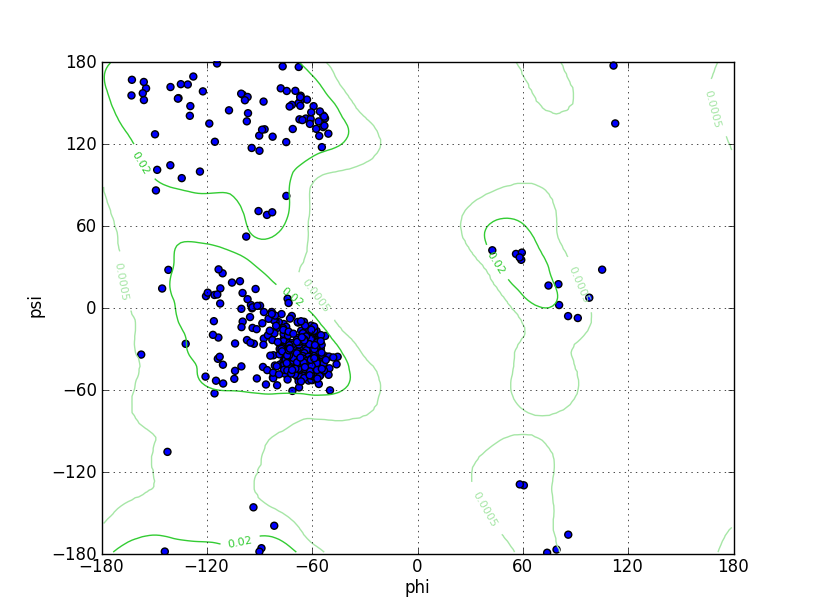
Ramz Z Score:-0.76 ±0.33 Residues in Favored Region:539 Ramachandran favored:98.72% Number of Outliers:0 Ramachandran outliers:0.00% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
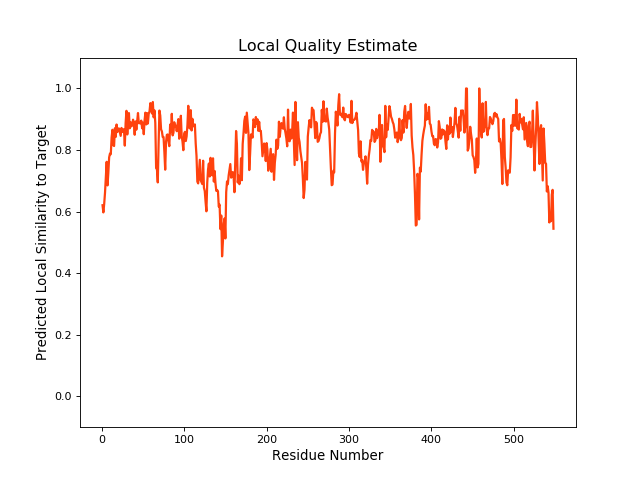
QMEANBrane Score:0.82 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Rattus norvegicus (Rat) | |||||
| Gene Name | Slc6a3 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:4XPB_A Sequence Length:549 Identity:56.364% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -1518.934 kcal/mol | |||
| Ramachandra Favored | Excellent |
||||
| QMEANBrane Quality | High |
||||
| Ramachandran Plot |
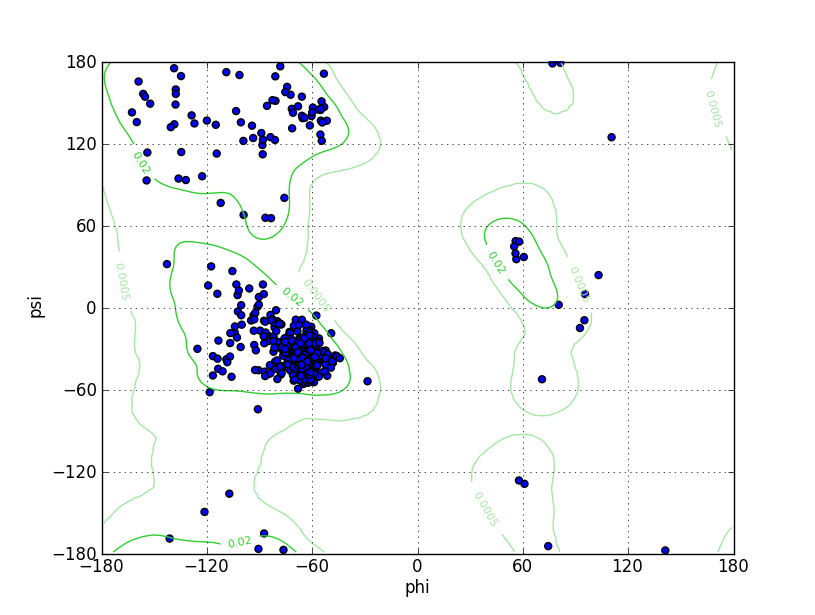
Ramz Z Score:-0.93 ±0.33 Residues in Favored Region:539 Ramachandran favored:98.54% Number of Outliers:0 Ramachandran outliers:0.00% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
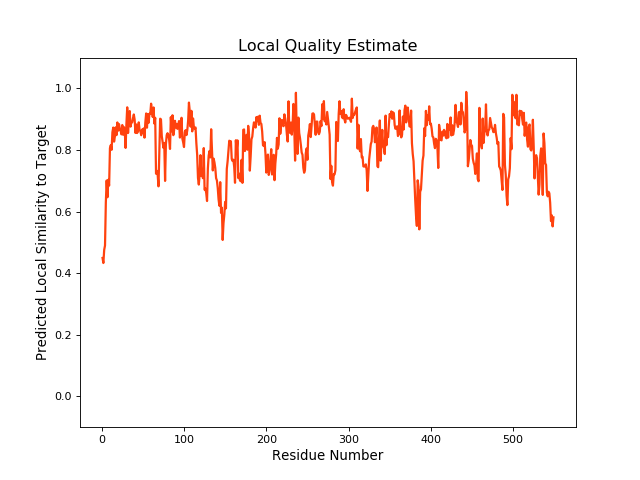
QMEANBrane Score:0.82 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Macaca fascicularis (Cynomolgus monkey) | |||||
| Gene Name | SLC6A3 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:4XPB_A Sequence Length:550 Identity:54.809% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -1516.618 kcal/mol | |||
| Ramachandra Favored | Excellent |
||||
| QMEANBrane Quality | High |
||||
| Ramachandran Plot |
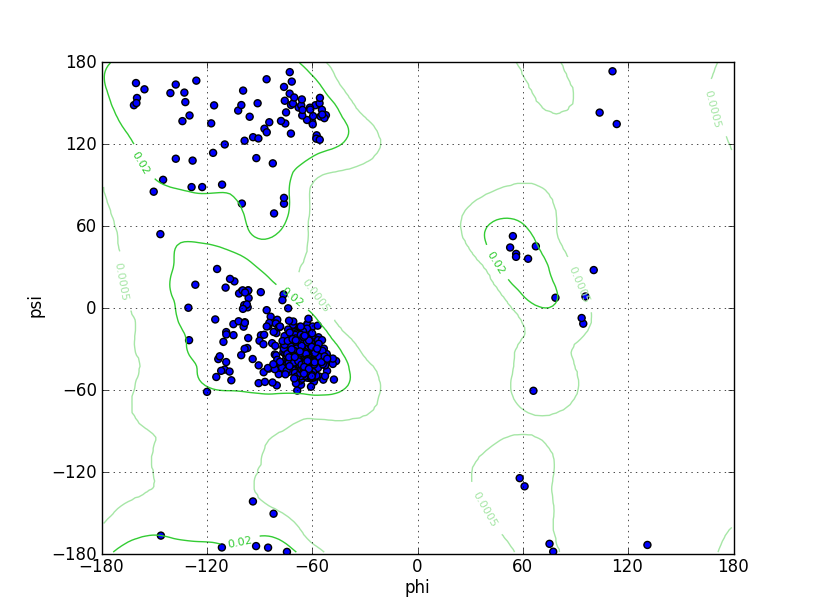
Ramz Z Score:-1.13 ±0.32 Residues in Favored Region:543 Ramachandran favored:99.09% Number of Outliers:0 Ramachandran outliers:0.00% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
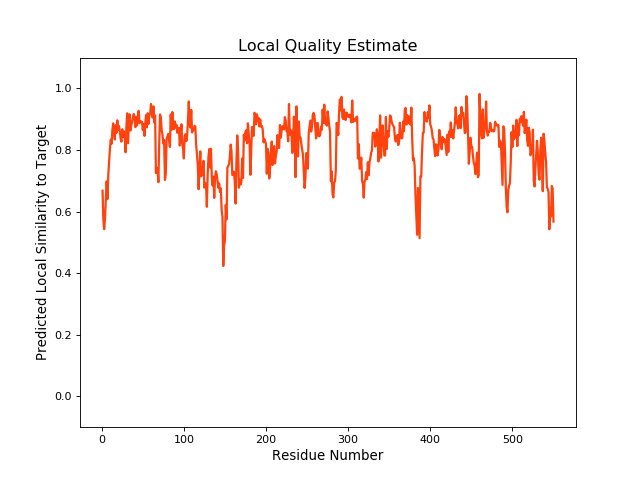
QMEANBrane Score:0.82 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| References | |||||
| 1 | Neurotransmitter and psychostimulant recognition by the dopamine transporter. Nature. 2015 May 21;521(7552):322-7. | ||||
| 2 | X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature. 2013 Nov 7;503(7474):85-90. | ||||
| 3 | X-ray structures of Drosophila dopamine transporter in complex with nisoxetine and reboxetine. Nat Struct Mol Biol. 2015 Jun;22(6):506-508. | ||||
| 4 | Structural basis of norepinephrine recognition and transport inhibition in neurotransmitter transporters. Nat Commun. 2021;12(1):2199. | ||||
| 5 | Structural insights into GABA transport inhibition using an engineered neurotransmitter transporter. EMBO J. 2022;41(15):e110735. | ||||
| 6 | Cryo-EM structure of the dopamine transporter with a novel atypical non-competitive inhibitor bound to the orthosteric site. J Neurochem. 2024;168(9):2043-2055. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.