Detail Information of DT Wildtype Structure
| General Information of Drug Transporter (DT) | ||||||
|---|---|---|---|---|---|---|
| DT ID | DTD0453 Transporter Info | |||||
| Gene Name | SLC6A20 | |||||
| Transporter Name | Sodium/imino-acid transporter 1 | |||||
| Gene ID | ||||||
| UniProt ID | ||||||
| Modelled Structures of This DT | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Detail Information of Structural Model | ||||||
| Method | Homology modeling | |||||
| Template PDB | 6M17_A | Identity | 46.01% | |||
| Performance | Minimized Score | -1377.55 kcal/mol | ||||
| Ramachandra Favored | Poor |
|||||
| QMEANBrane Quality | High |
|||||
| Ramachandran Plot |
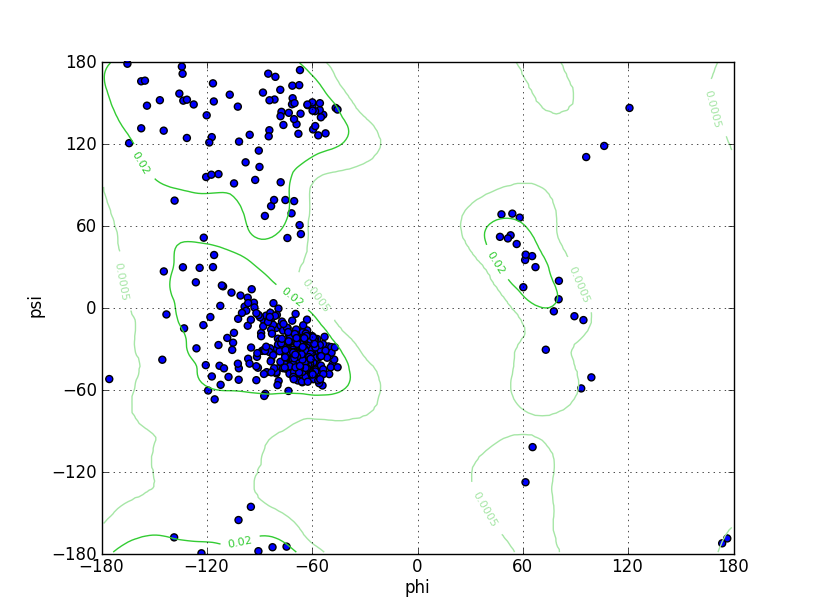
|
|||||
| Click to Save Ramachandran Plot in PNG Format | ||||||
| Ramz Z Score | -1.64 ±0.3 | |||||
| Residues in Favored Region | 553 | Ramachandran favored | 94.85% | |||
| Number of Outliers | 5 | Ramachandran outliers | 0.86% | |||
| Local Quality |

|
|||||
| Click to Save Local Quality Plot in PNG Format | ||||||
| QMEANBrane Score | 0.81 | |||||
| AlphaFold Protein Structure Database | ||||||
| SWISS-MODEL Database | ||||||
| Experimental Structures of This DT | ||||||
| PDB ID | Scanning Method | Resolution | Expression System | Details | Ref | |
| 7Y75 | EM | 3.1 Å | Homo sapiens | [1] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | B/D | |||||
| Sequence Length | 613 | Mutation | No | |||
| 7Y76 | EM | 3.2 Å | Homo sapiens | [1] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | B/D | |||||
| Sequence Length | 613 | Mutation | No | |||
| 8I91 | EM | 3.3 Å | Homo sapiens | [2] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | B/D | |||||
| Sequence Length | 613 | Mutation | No | |||
| 8P2W | EM | 3.76 Å | Homo sapiens | [3] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | C | |||||
| Sequence Length | 641 | Mutation | No | |||
| 8P2X | EM | 3.59 Å | Homo sapiens | [3] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | C/D | |||||
| Sequence Length | 641 | Mutation | No | |||
| 8P2Y | EM | 3.46 Å | Homo sapiens | [3] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | C/D | |||||
| Sequence Length | 641 | Mutation | No | |||
| 8P2Z | EM | 3.5 Å | Homo sapiens | [3] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | C | |||||
| Sequence Length | 641 | Mutation | No | |||
| 8P30 | EM | 3.29 Å | Homo sapiens | [3] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | C/D | |||||
| Sequence Length | 641 | Mutation | No | |||
| References | ||||||
| 1 | Structures of ACE2-SIT1 recognized by Omicron variants of SARS-CoV-2. Cell Discov. 2022;8(1):123. | |||||
| 2 | Structural insight into the substrate recognition and transport mechanism of amino acid transporter complex ACE2-B(0)AT1 and ACE2-SIT1. Cell Discov. 2023;9(1):93. | |||||
| 3 | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2. Nat Commun. 2024;15(1):5503. | |||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
