Detail Information of DT Wildtype Structure
| General Information of Drug Transporter (DT) | ||||||
|---|---|---|---|---|---|---|
| DT ID | DTD0422 Transporter Info | |||||
| Gene Name | SLC5A2 | |||||
| Transporter Name | Sodium/glucose cotransporter 2 | |||||
| Gene ID | ||||||
| UniProt ID | ||||||
| Modelled Structures of This DT | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Detail Information of Structural Model | ||||||
| Method | Homology modeling | |||||
| Template PDB | 2XQ2_A | Identity | 32.842% | |||
| Performance | Minimized Score | -979.24 kcal/mol | ||||
| Ramachandra Favored | Poor |
|||||
| QMEANBrane Quality | Medium |
|||||
| Ramachandran Plot |
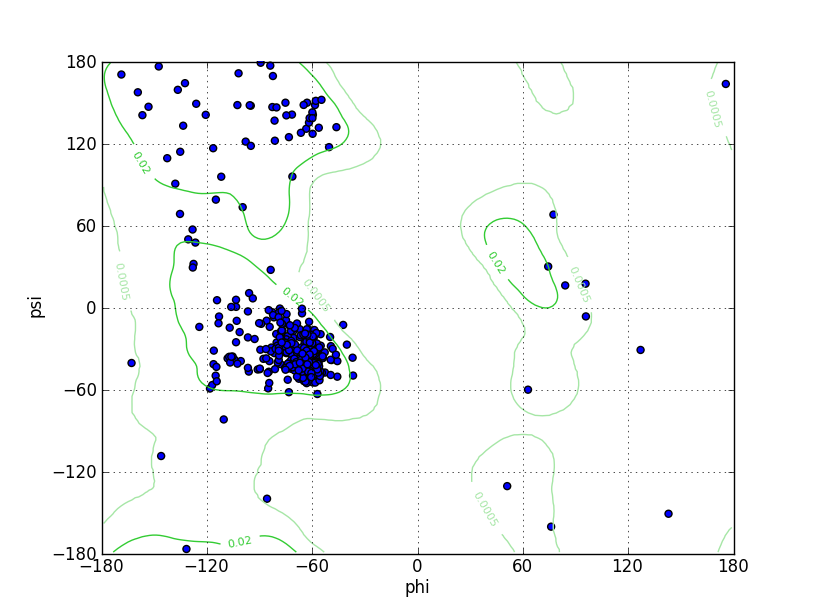
|
|||||
| Click to Save Ramachandran Plot in PNG Format | ||||||
| Ramz Z Score | -1.33 ±0.35 | |||||
| Residues in Favored Region | 441 | Ramachandran favored | 94.43% | |||
| Number of Outliers | 5 | Ramachandran outliers | 1.07% | |||
| Local Quality |
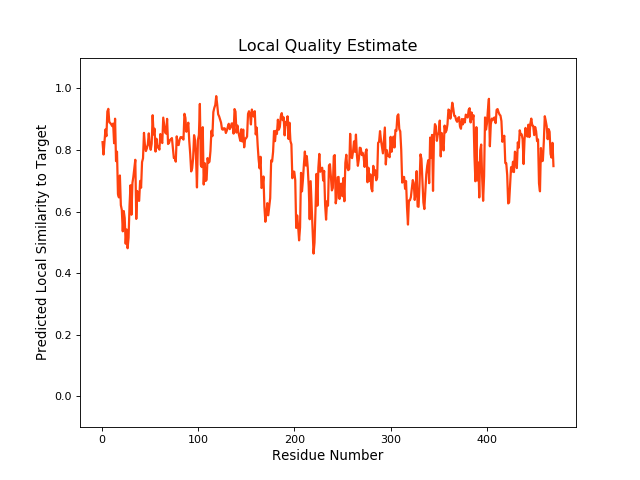
|
|||||
| Click to Save Local Quality Plot in PNG Format | ||||||
| QMEANBrane Score | 0.79 | |||||
| AlphaFold Protein Structure Database | ||||||
| SWISS-MODEL Database | ||||||
| Experimental Structures of This DT | ||||||
| PDB ID | Scanning Method | Resolution | Expression System | Details | Ref | |
| 7VSI | EM | 2.95 Å | Homo sapiens | [1] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | A | |||||
| Sequence Length | 672 | Mutation | No | |||
| 7YNJ | EM | 3.33 Å | Homo sapiens | [2] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | A | |||||
| Sequence Length | 672 | Mutation | No | |||
| 7YNK | EM | 3.48 Å | Homo sapiens | [2] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | A | |||||
| Sequence Length | 672 | Mutation | No | |||
| 8HB0 | EM | 2.9 Å | Homo sapiens | [3] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | A | |||||
| Sequence Length | 676 | Mutation | No | |||
| 8HDH | EM | 3.1 Å | Homo sapiens | [3] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | A | |||||
| Sequence Length | 676 | Mutation | No | |||
| 8HEZ | EM | 2.8 Å | Homo sapiens | [3] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | A | |||||
| Sequence Length | 676 | Mutation | No | |||
| 8HG7 | EM | 3.1 Å | Homo sapiens | [3] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | A | |||||
| Sequence Length | 676 | Mutation | No | |||
| 8HIN | EM | 3.3 Å | Homo sapiens | [3] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | A | |||||
| Sequence Length | 676 | Mutation | No | |||
| References | ||||||
| 1 | Structural basis of inhibition of the human SGLT2-MAP17 glucose transporter. Nature. 2022;601(7892):280-284. | |||||
| 2 | Structures of human SGLT in the occluded state reveal conformational changes during sugar transport. Nat Commun. 2023;14(1):2920. | |||||
| 3 | Transport and inhibition mechanism of the human SGLT2-MAP17 glucose transporter. Nat Struct Mol Biol. 2024;31(1):159-169. | |||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
