Detail Information of Mutation-induced Spatial Variation
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0015 Transporter Info | ||||
| Gene Name | ABCC4 | ||||
| Transporter Name | Multidrug resistance-associated protein 4 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Mutation-induced Structural Variation (MSV) of This DT | |||||
| PDB ID | |||||
| Expression System | Homo sapiens | ||||
| Method | EM | ||||
| Resolution | 3.2 Å | ||||
| Corresponding chain | A | ||||
| Mutation Site(s) | E1202Q | [1] | |||
| Structure Details | |||||
| Modeling Method | EM | ||||
| Publication year | 2023 | ||||
| PDB ID | Scanning Method | Resolution | Mutation Sites | Details | Ref |
| 8BWO | EM | 3.2 Å | E1202Q | [1] | |
| Structure | |||||
| Expression System | Homo sapiens | ||||
| Corresponding chain | A | ||||
| Sequence Length | 1-1325 | Mutation | Yes | ||
| Mutation-induced Spatial Variation (MSV) of This DT | |||||
| SNP ID | |||||
| Variation Type | G/W | ||||
| Site of GPD | chr13:95210754 (GRCh38.p12) | ||||
| Allele(s) in dbSNP | C>A | ||||
| Minor Allele Frequency | A=0.0517/259 | ||||
| GPD Details | GPD Info | ||||
| Structure Details | |||||
| Modeling Method | Homology modeling | ||||
| Template PDB | 5UJ9_A | Identity | 36.634% | ||
| Model Performance | Minimized Score | -2124.76 kcal/mol | |||
| Ramachandra Favored | Poor |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |

|
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Ramz Z Score | -1.99 ±0.21 | ||||
| Residues in Favored Region | 1191 | Percentage of Favorable Residues | 94.08% | ||
| Number of Outliers | 24 | Ramachandran outliers | 1.90% | ||
| Local Quality |

|
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| QMEANBrane Score | 0.72 | ||||
| SNP ID | |||||
| Variation Type | K/N | ||||
| Site of GPD | chr13:95062722 (GRCh38.p12) | ||||
| Allele(s) in dbSNP | C>G / C>T | ||||
| Minor Allele Frequency | C=0.2057/1030 | ||||
| GPD Details | GPD Info | ||||
| Structure Details | |||||
| Modeling Method | Homology modeling | ||||
| Template PDB | 5UJ9_A | Identity | 36.56% | ||
| Model Performance | Minimized Score | -2085.818 kcal/mol | |||
| Ramachandra Favored | Poor |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |

|
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Ramz Z Score | -1.93 ±0.21 | ||||
| Residues in Favored Region | 1187 | Percentage of Favorable Residues | 93.76% | ||
| Number of Outliers | 25 | Ramachandran outliers | 1.97% | ||
| Local Quality |
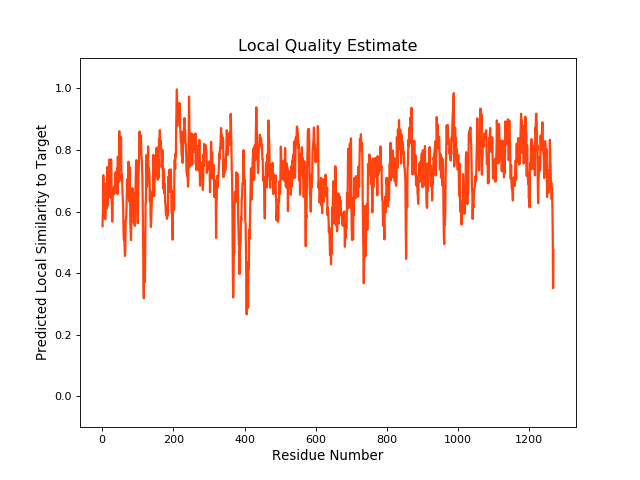
|
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| QMEANBrane Score | 0.72 | ||||
| SNP ID | |||||
| Variation Type | E/K | ||||
| Site of GPD | chr13:95163161 (GRCh38.p12) | ||||
| Allele(s) in dbSNP | C>T | ||||
| Minor Allele Frequency | T=0.0312/156 | ||||
| GPD Details | GPD Info | ||||
| Structure Details | |||||
| Modeling Method | Homology modeling | ||||
| Template PDB | 5UJ9_A | Identity | 36.634% | ||
| Model Performance | Minimized Score | -2141.226 kcal/mol | |||
| Ramachandra Favored | Poor |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
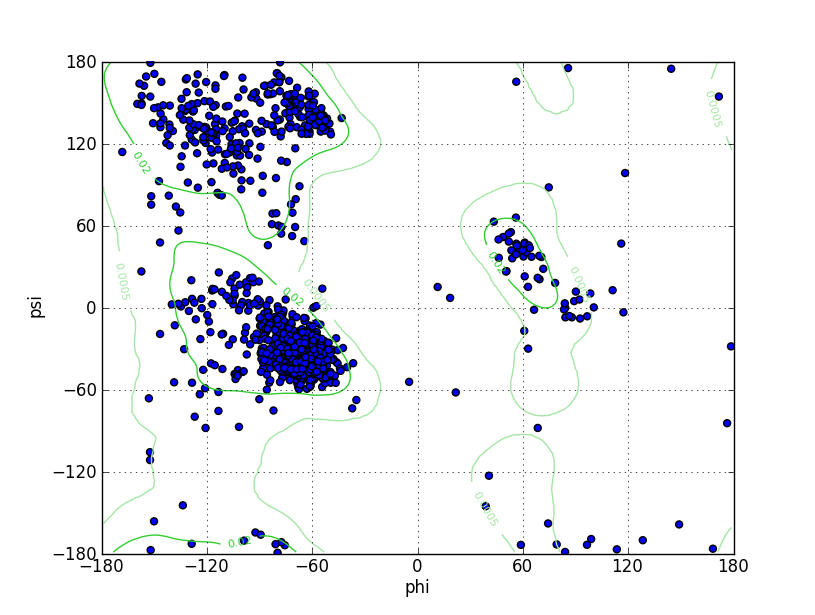
|
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Ramz Z Score | -2.02 ±0.21 | ||||
| Residues in Favored Region | 1191 | Percentage of Favorable Residues | 94.08% | ||
| Number of Outliers | 23 | Ramachandran outliers | 1.82% | ||
| Local Quality |

|
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| QMEANBrane Score | 0.73 | ||||
| PDB ID | Scanning Method | Resolution | Mutation Sites | Details | Ref |
| 8BWO | EM | 3.2 Å | E1202Q | [1] | |
| Structure | |||||
| Expression System | Homo sapiens | ||||
| Corresponding chain | A | ||||
| Sequence Length | 1-1325 | Mutation | Yes | ||
| References | |||||
| 1 | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4. Structure. 2023;31(11):1407-1418.e6. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
