Modelled Stucture
Method:Homology modeling
Teplate PDB:4XE5_A
Identity:64.902%
Minimized Score:-2634.284
Detail: Structure Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0540 Transporter Info | ||||
| Gene Name | ATP12A | ||||
| Protein Name | Potassium-transporting ATPase alpha chain 2 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Modelled Stucture Method:Homology modeling Teplate PDB:4XE5_A Identity:64.902% Minimized Score:-2634.284 Detail: Structure Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Mus musculus (Mouse) | |||||
| Gene Name | Atp12a | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5YLU_A Sequence Length:1030 Identity:63.988% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -2894.548 kcal/mol | |||
| Ramachandra Favored | Excellent |
||||
| QMEANBrane Quality | High |
||||
| Ramachandran Plot |
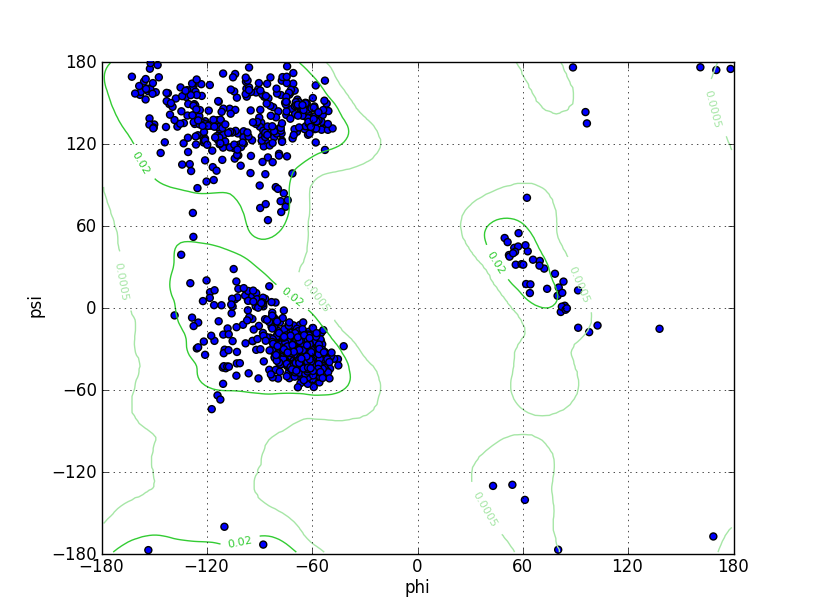
Ramz Z Score:-0.28 ±0.25 Residues in Favored Region:1016 Ramachandran favored:98.83% Number of Outliers:1 Ramachandran outliers:0.10% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
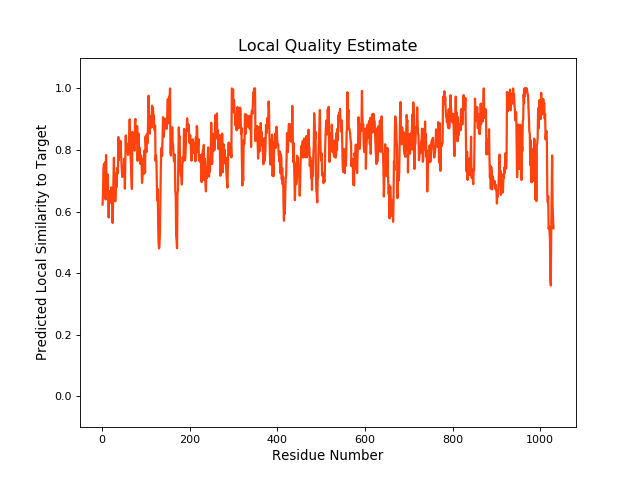
QMEANBrane Score:0.81 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Rattus norvegicus (Rat) | |||||
| Gene Name | Atp12a | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5YLU_A Sequence Length:991 Identity:65.323% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -2928.896 kcal/mol | |||
| Ramachandra Favored | Excellent |
||||
| QMEANBrane Quality | High |
||||
| Ramachandran Plot |
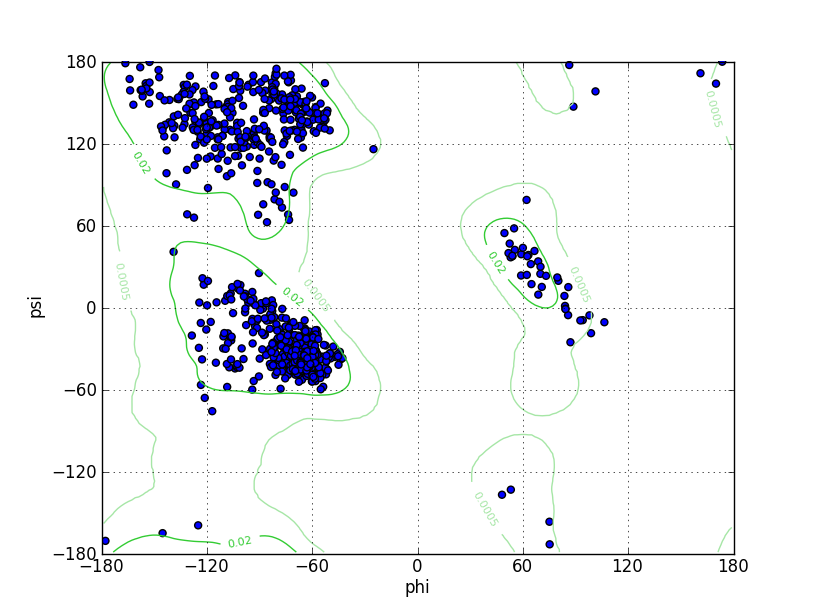
Ramz Z Score:-0.02 ±0.25 Residues in Favored Region:972 Ramachandran favored:98.28% Number of Outliers:1 Ramachandran outliers:0.10% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
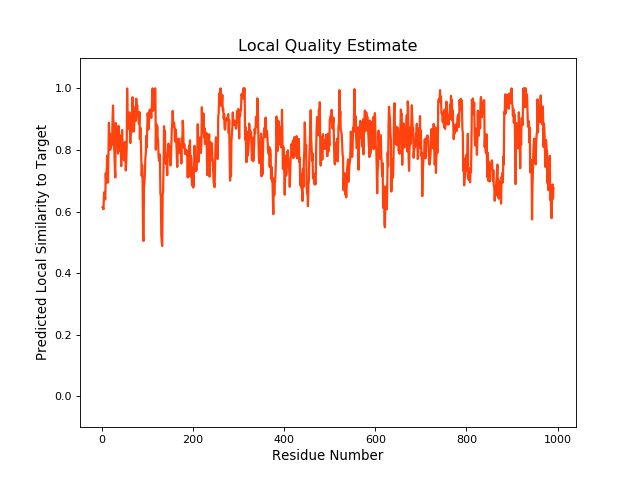
QMEANBrane Score:0.81 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Oryctolagus cuniculus (Rabbit) | |||||
| Gene Name | ATP12A | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5YLU_A Sequence Length:1027 Identity:64.07% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -2915.629 kcal/mol | |||
| Ramachandra Favored | Excellent |
||||
| QMEANBrane Quality | High |
||||
| Ramachandran Plot |
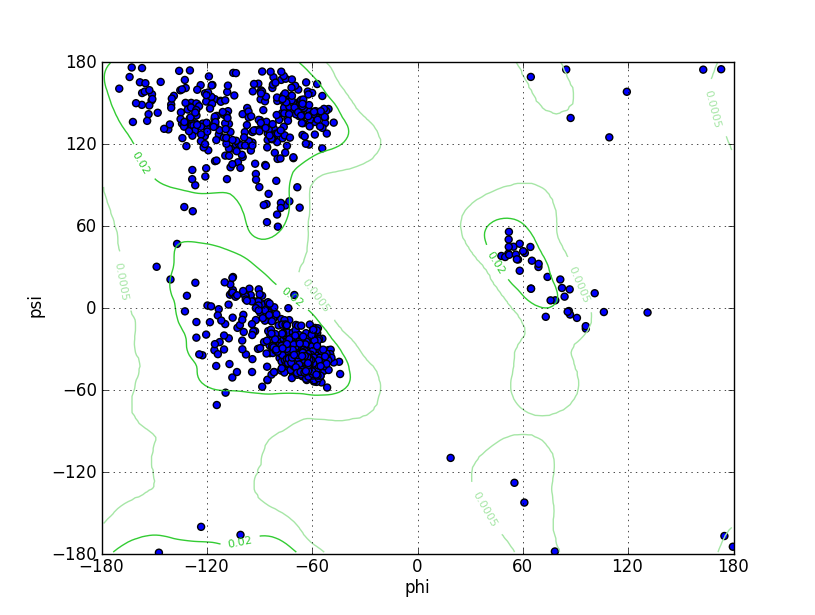
Ramz Z Score:-0.09 ±0.24 Residues in Favored Region:1013 Ramachandran favored:98.83% Number of Outliers:1 Ramachandran outliers:0.10% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
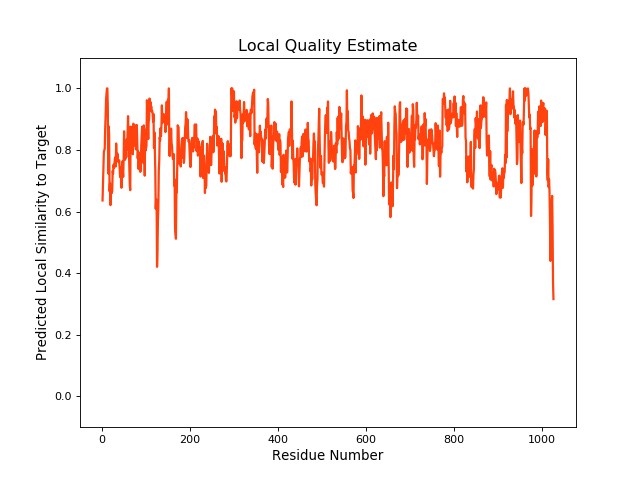
QMEANBrane Score:0.81 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| References | |||||
| 1 | Structure and function of H(+)/K(+) pump mutants reveal Na(+)/K(+) pump mechanisms. Nat Commun. 2022;13(1):5270. | ||||
| 2 | An unusual conformation from Na(+)-sensitive non-gastric proton pump mutants reveals molecular mechanisms of cooperative Na(+)-binding. Biochim Biophys Acta Mol Cell Res. 2023;1870(7):119543. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.