Crystal Structure of This DT
Crystallisation Method:EM
Corresponding Chain:A/B/C/D
Sequence Length:2-208
Detail: Struture Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0523 Transporter Info | ||||
| Gene Name | CACNG2 | ||||
| Protein Name | Voltage-dependent calcium channel gamma-2 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Crystal Structure of This DT Crystallisation Method:EM Corresponding Chain:A/B/C/D Sequence Length:2-208 Detail: Struture Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Mus musculus (Mouse) | |||||
| Gene Name | Cacng2 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5KK2_E Sequence Length:323 Identity:100% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -499.438 kcal/mol | |||
| Ramachandra Favored | Medium |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |

Ramz Z Score:-1.42 ±0.42 Residues in Favored Region:305 Ramachandran favored:95.02% Number of Outliers:1 Ramachandran outliers:0.31% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
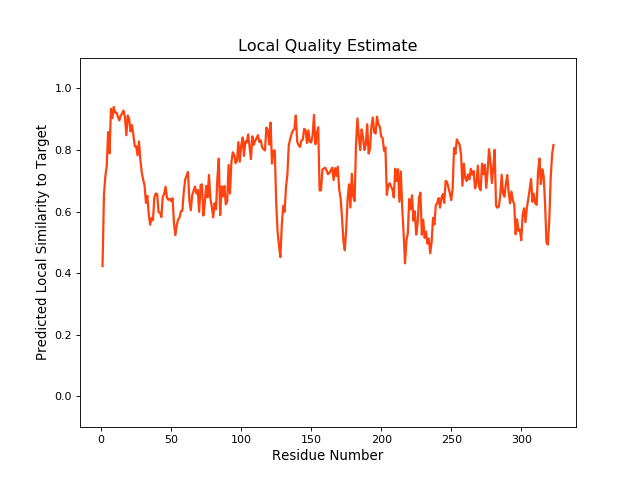
QMEANBrane Score:0.7 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Experimental Structures of This Model Oganism | |||||
| PDB ID | Scanning Method | Resolution | Expression System | Details | Ref |
| 3JXT | X-ray | 1.5 Å | Escherichia coli | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | C/D | ||||
| Sequence Length | 318-323 | Mutation | Yes | ||
| 4X3H | X-ray | 2.4 Å | Escherichia coli | [ 2] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | B | ||||
| Sequence Length | 225-233 | Mutation | No | ||
| 5KBS | EM | 8.7 Å | Homo sapiens | [ 3] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B/C/D | ||||
| Sequence Length | 2-208 | Mutation | Yes | ||
| 5KBT | EM | 6.4 Å | Homo sapiens | [ 3] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B/C/D | ||||
| Sequence Length | 2-208 | Mutation | Yes | ||
| 5KBU | EM | 7.8 Å | Homo sapiens | [ 3] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B/C/D | ||||
| Sequence Length | 2-208 | Mutation | Yes | ||
| 5WEO | EM | 4.2 Å | Homo sapiens | [ 4] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B/C/D | ||||
| Sequence Length | 2-208 | Mutation | Yes | ||
| 8FP4 | EM | 2.4 Å | Homo sapiens | [ 5] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | E/F/G/H | ||||
| Sequence Length | 1-323 | Mutation | No | ||
| 8FP9 | EM | 2.44 Å | Homo sapiens | [ 5] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | E/F/G/H | ||||
| Sequence Length | 1-323 | Mutation | No | ||
| Rattus norvegicus (Rat) | |||||
| Gene Name | Cacng2 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure | |||||
| Click to Save PDB File in PDB Format | |||||
| Experimental Structures of This Model Oganism | |||||
| PDB ID | Scanning Method | Resolution | Expression System | Details | Ref |
| References | |||||
| 1 | Caged mono- and divalent ligands for light-assisted disruption of PDZ domain-mediated interactions. J Am Chem Soc. 2013 Mar 27;135(12):4580-3. | ||||
| 2 | Structural basis of arc binding to synaptic proteins: implications for cognitive disease. Neuron. 2015 Apr 22;86(2):490-500. | ||||
| 3 | Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy. Science. 2016 Jul 1;353(6294):83-6. | ||||
| 4 | Channel opening and gating mechanism in AMPA-subtype glutamate receptors. Nature. 2017 Sep 7;549(7670):60-65. | ||||
| 5 | The open gate of the AMPA receptor forms a Ca(2+) binding site critical in regulating ion transport. Nat Struct Mol Biol. 2024;31(4):688-700. | ||||
| 6 | Proton-triggered rearrangement of the AMPA receptor N-terminal domains impacts receptor kinetics and synaptic localization. Nat Struct Mol Biol. 2024;31(10):1601-1613. | ||||
| 7 | Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM. Cell. 2017 Sep 7;170(6):1234-1246.e14. | ||||
| 8 | Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM. Nature. 2016 Aug 4;536(7614):108-11. | ||||
| 9 | Architecture and subunit arrangement of native AMPA receptors elucidated by cryo-EM. Science. 2019 Apr 26;364(6438):355-362. | ||||
| 10 | Opening of glutamate receptor channel to subconductance levels. Nature. 2022;605(7908):172-178. | ||||
| 11 | Structural mobility tunes signalling of the GluA1 AMPA glutamate receptor. Nature. 2023;621(7980):877-882. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.