Crystal Structure of This DT
Crystallisation Method:EM
Corresponding Chain:A/B/C/D
Sequence Length:66-1179
Detail: Struture Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0512 Transporter Info | ||||
| Gene Name | KCNMA1 | ||||
| Protein Name | Calcium-activated potassium channel subunit alpha-1 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Crystal Structure of This DT Crystallisation Method:EM Corresponding Chain:A/B/C/D Sequence Length:66-1179 Detail: Struture Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Bos taurus (Bovine) | |||||
| Gene Name | KCNMA1 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:6V22_A Sequence Length:1059 Identity:99.528% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -2606.632 kcal/mol | |||
| Ramachandra Favored | Medium |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
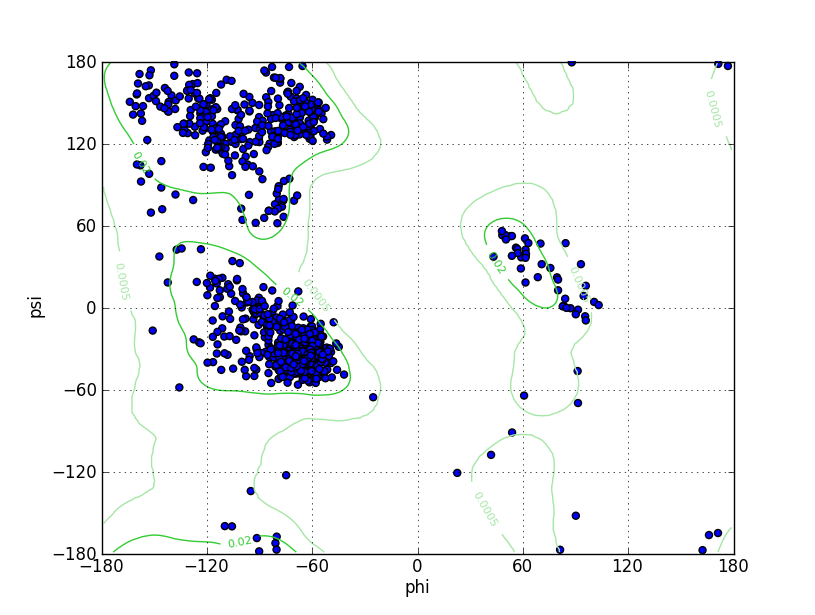
Ramz Z Score:-1.1 ±0.24 Residues in Favored Region:1028 Ramachandran favored:97.26% Number of Outliers:7 Ramachandran outliers:0.66% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
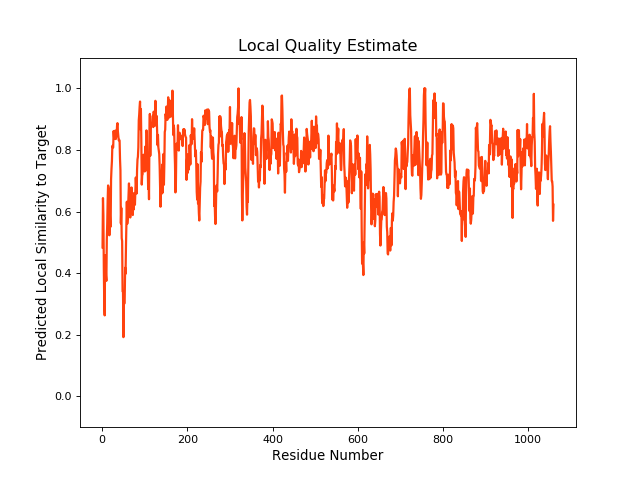
QMEANBrane Score:0.76 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Macaca mulatta (Rhesus macaque) | |||||
| Gene Name | KCNMA1 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:6V22_A Sequence Length:1059 Identity:99.717% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -2618.645 kcal/mol | |||
| Ramachandra Favored | Medium |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
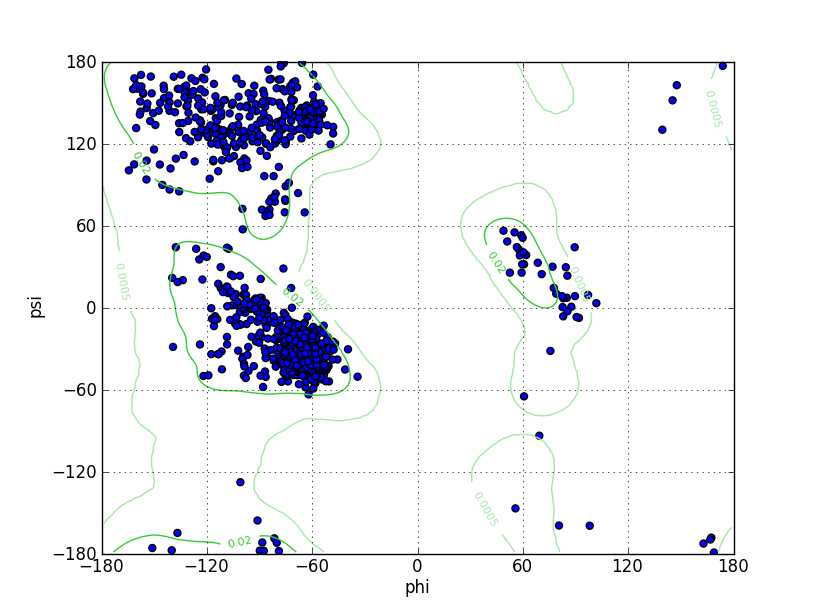
Ramz Z Score:-0.72 ±0.25 Residues in Favored Region:1034 Ramachandran favored:97.82% Number of Outliers:3 Ramachandran outliers:0.28% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
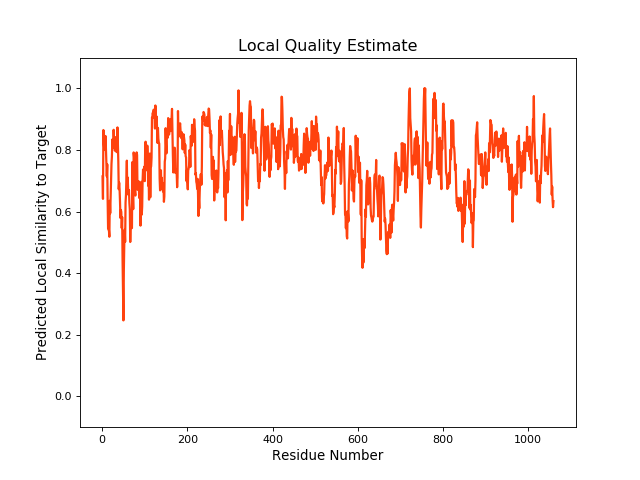
QMEANBrane Score:0.76 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Xenopus laevis (African clawed frog) | |||||
| Gene Name | kcnma1 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:6V22_A Sequence Length:1059 Identity:92.925% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -2647.239 kcal/mol | |||
| Ramachandra Favored | Medium |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
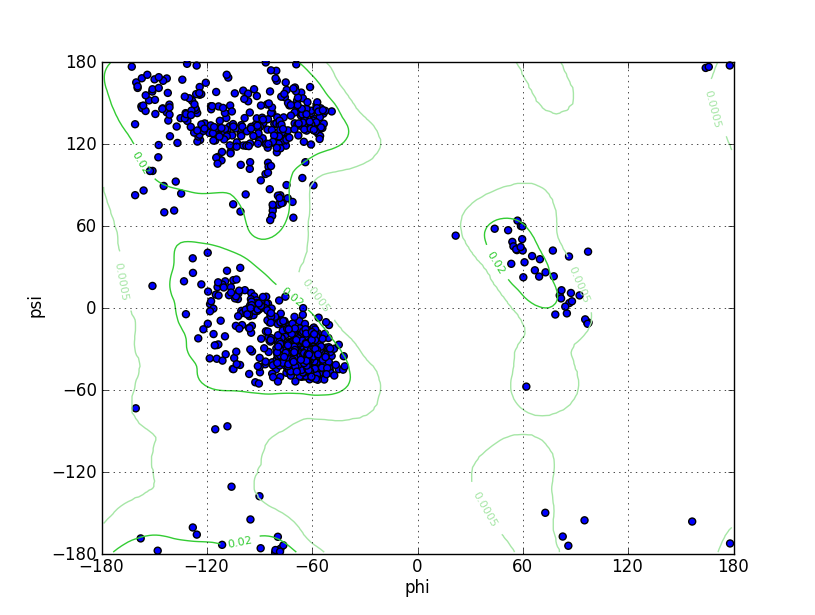
Ramz Z Score:0.13 ±0.25 Residues in Favored Region:1028 Ramachandran favored:97.26% Number of Outliers:3 Ramachandran outliers:0.28% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
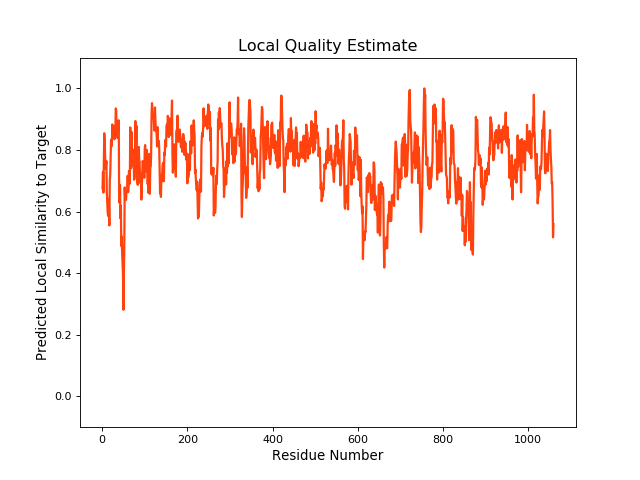
QMEANBrane Score:0.76 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| References | |||||
| 1 | Cryo-EM structure of the Slo1 potassium channel with the auxiliary 1 subunit suggests a mechanism for depolarization-independent activation. FEBS Lett. 2024;598(8):875-888. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.