Modelled Stucture
Method:Homology modeling
Teplate PDB:2XUT_A
Identity:30.7%
Minimized Score:-615.174
Detail: Structure Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0022 Transporter Info | ||||
| Gene Name | SLC15A2 | ||||
| Protein Name | Peptide transporter 2 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Modelled Stucture Method:Homology modeling Teplate PDB:2XUT_A Identity:30.7% Minimized Score:-615.174 Detail: Structure Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Mus musculus (Mouse) | |||||
| Gene Name | Slc15a2 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:2XUT_A Sequence Length:391 Identity:32.5% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -610.124 kcal/mol | |||
| Ramachandra Favored | Poor |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
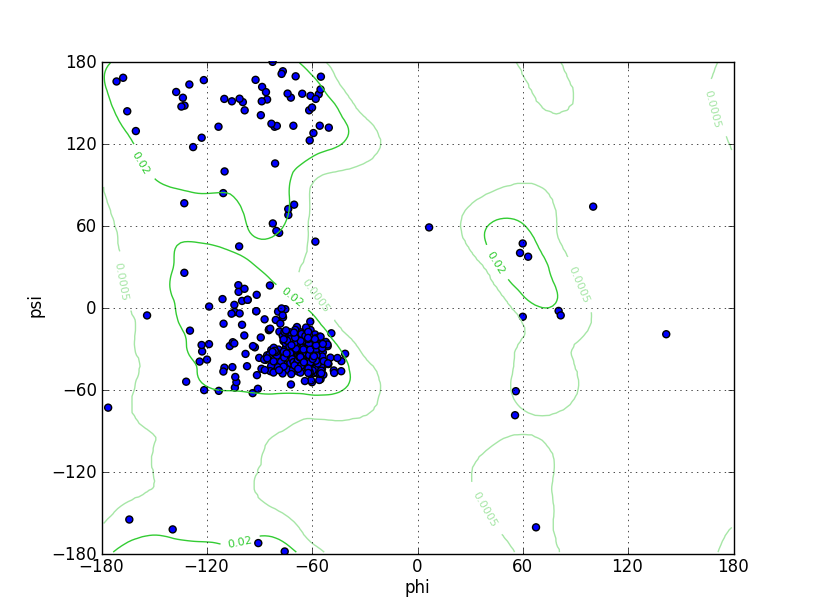
Ramz Z Score:-2.88 ±0.39 Residues in Favored Region:363 Ramachandran favored:93.32% Number of Outliers:8 Ramachandran outliers:2.06% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
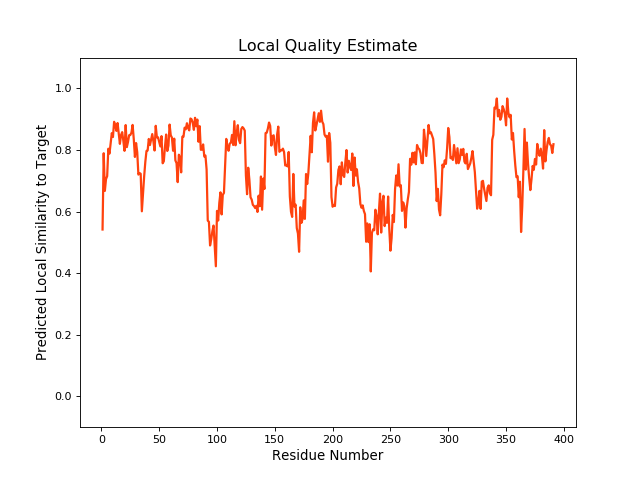
QMEANBrane Score:0.74 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Rattus norvegicus (Rat) | |||||
| Gene Name | Slc15a2 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure | |||||
| Click to Save PDB File in PDB Format | |||||
| Experimental Structures of This Model Oganism | |||||
| PDB ID | Scanning Method | Resolution | Expression System | Details | Ref |
| 5A9I | X-ray | 2.84 Å | Escherichia coli | [ 1] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A/B/C | ||||
| Sequence Length | 410-601 | Mutation | No | ||
| 7NQK | EM | 3.5 Å | Homo sapiens | [ 2] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 1-729 | Mutation | No | ||
| 9BIR | EM | 3.1 Å | Homo sapiens | [ 3] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 1-729 | Mutation | No | ||
| 9BIS | EM | 3.2 Å | Homo sapiens | [ 3] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 1-729 | Mutation | No | ||
| 9BIT | EM | 3.1 Å | Homo sapiens | [ 3] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 1-729 | Mutation | No | ||
| 9BIU | EM | 2.9 Å | Homo sapiens | [ 3] | |
| Structure | |||||
| Click to Save PDB File in TXT Format | |||||
| Corresponding chain | A | ||||
| Sequence Length | 1-729 | Mutation | No | ||
| Oryctolagus cuniculus (Rabbit) | |||||
| Gene Name | SLC15A2 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:2XUT_A Sequence Length:362 Identity:31.618% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -529.326 kcal/mol | |||
| Ramachandra Favored | Poor |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
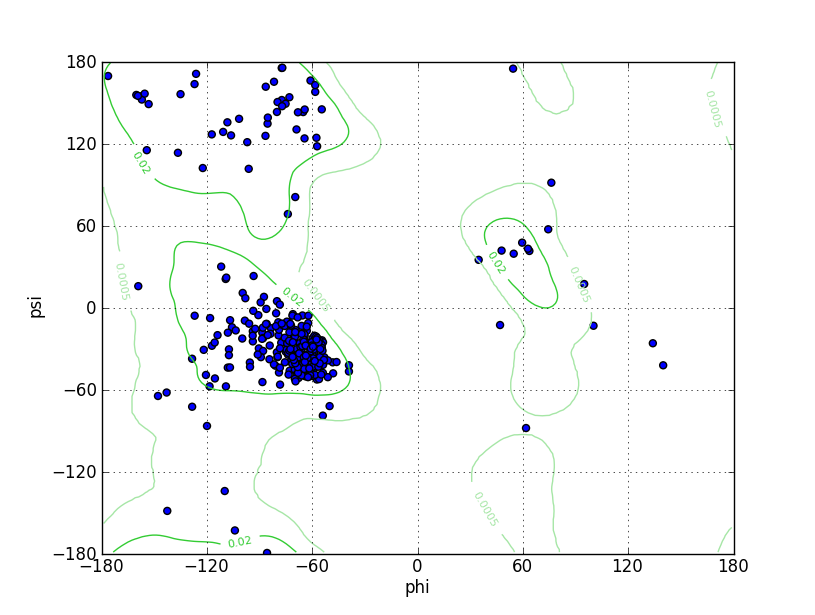
Ramz Z Score:-2.46 ±0.38 Residues in Favored Region:337 Ramachandran favored:93.61% Number of Outliers:8 Ramachandran outliers:2.22% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
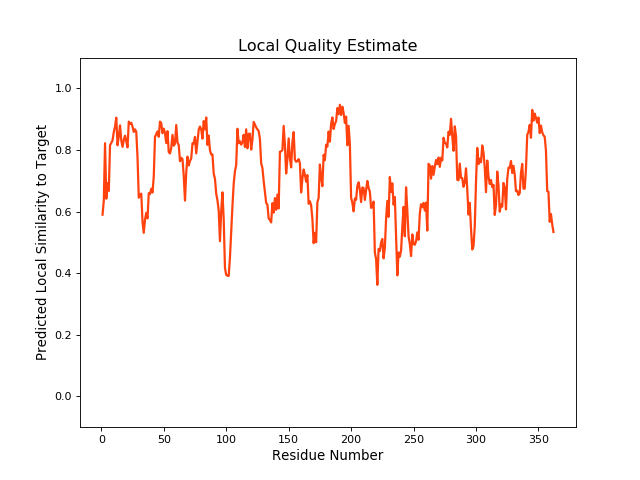
QMEANBrane Score:0.74 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| References | |||||
| 1 | Crystal Structures of the Extracellular Domain from PepT1 and PepT2 Provide Novel Insights into Mammalian Peptide Transport. Structure. 2015 Oct 6;23(10):1889-1899. | ||||
| 2 | Cryo-EM structure of PepT2 reveals structural basis for proton-coupled peptide and prodrug transport in mammals. Sci Adv. 2021;7(35). | ||||
| 3 | Structural basis for antibiotic transport and inhibition in PepT2. Nat Commun. 2024;15(1):8755. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.