Modelled Stucture
Method:Homology modeling
Teplate PDB:6UY0_A
Identity:90.725%
Minimized Score:-3891.081
Detail: Structure Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0001 Transporter Info | ||||
| Gene Name | ABCC1 | ||||
| Protein Name | Multidrug resistance-associated protein 1 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Modelled Stucture Method:Homology modeling Teplate PDB:6UY0_A Identity:90.725% Minimized Score:-3891.081 Detail: Structure Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Mus musculus (Mouse) | |||||
| Gene Name | Abcc1 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:6UY0_A Sequence Length:1528 Identity:87.924% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -3572.937 kcal/mol | |||
| Ramachandra Favored | Medium |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
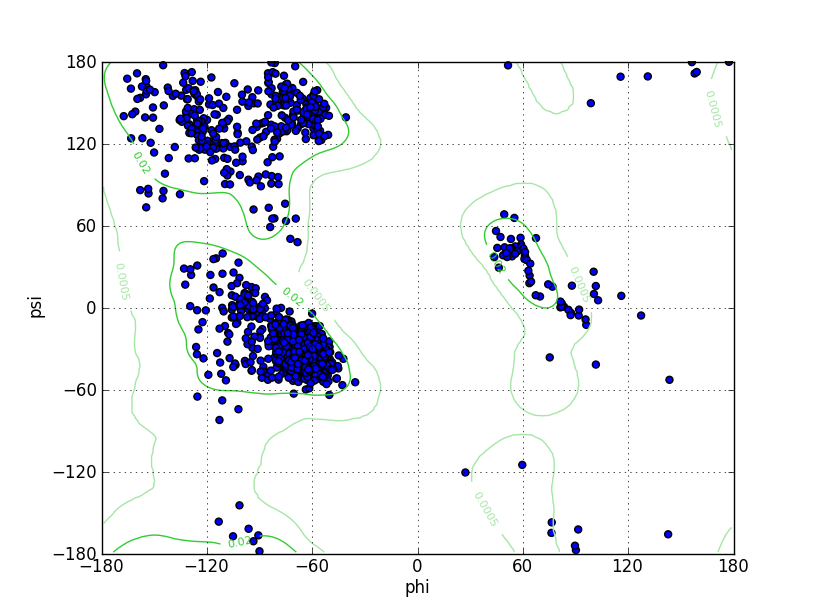
Ramz Z Score:-0.87 ±0.19 Residues in Favored Region:1490 Ramachandran favored:97.64% Number of Outliers:5 Ramachandran outliers:0.33% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
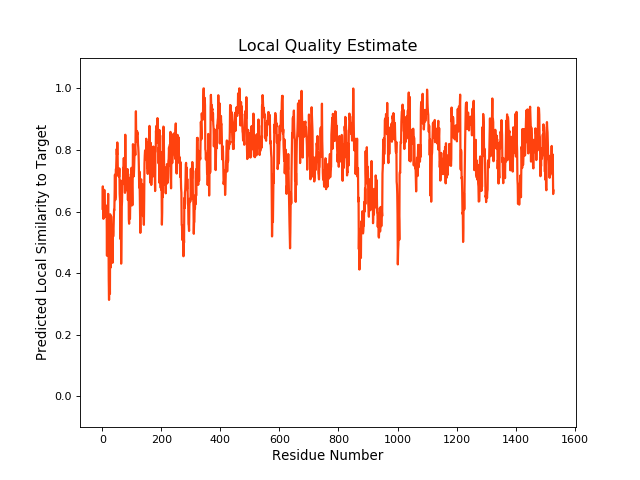
QMEANBrane Score:0.78 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Rattus norvegicus (Rat) | |||||
| Gene Name | Abcc1 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:6UY0_A Sequence Length:1532 Identity:87.859% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -3551.214 kcal/mol | |||
| Ramachandra Favored | Medium |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
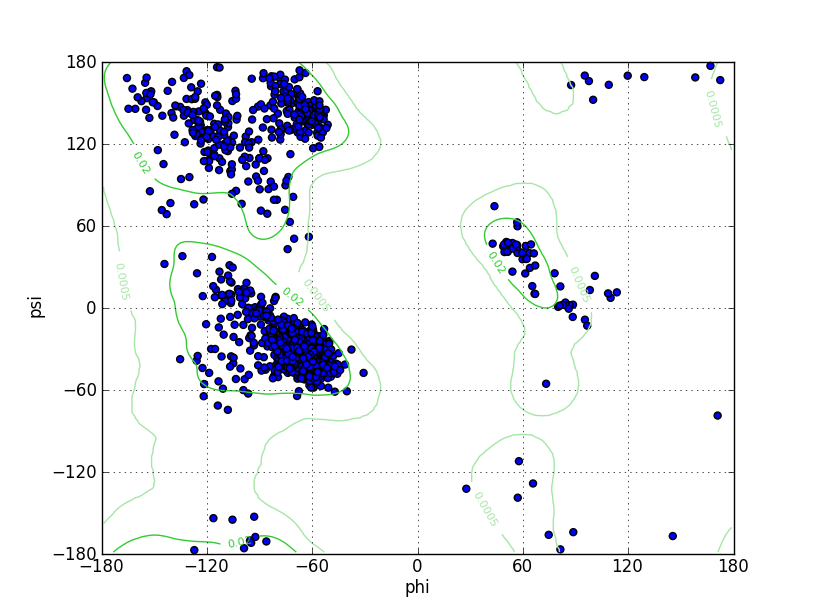
Ramz Z Score:-0.48 ±0.2 Residues in Favored Region:1492 Ramachandran favored:97.52% Number of Outliers:5 Ramachandran outliers:0.33% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
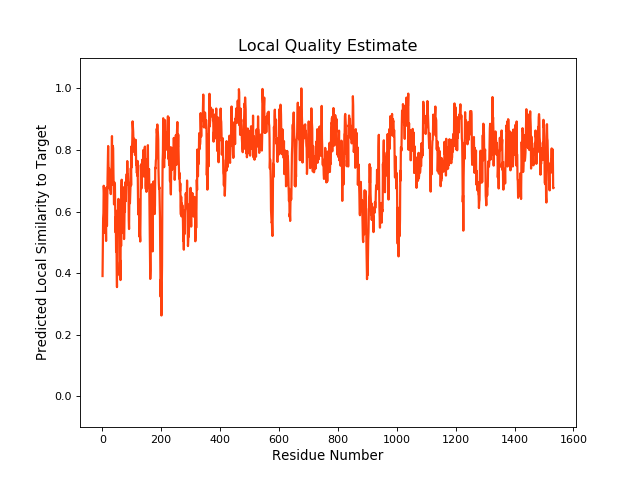
QMEANBrane Score:0.78 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Canis lupus familiaris (Dog) | |||||
| Gene Name | ABCC1 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:6UY0_A Sequence Length:1531 Identity:93.076% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -3490.303 kcal/mol | |||
| Ramachandra Favored | Excellent |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
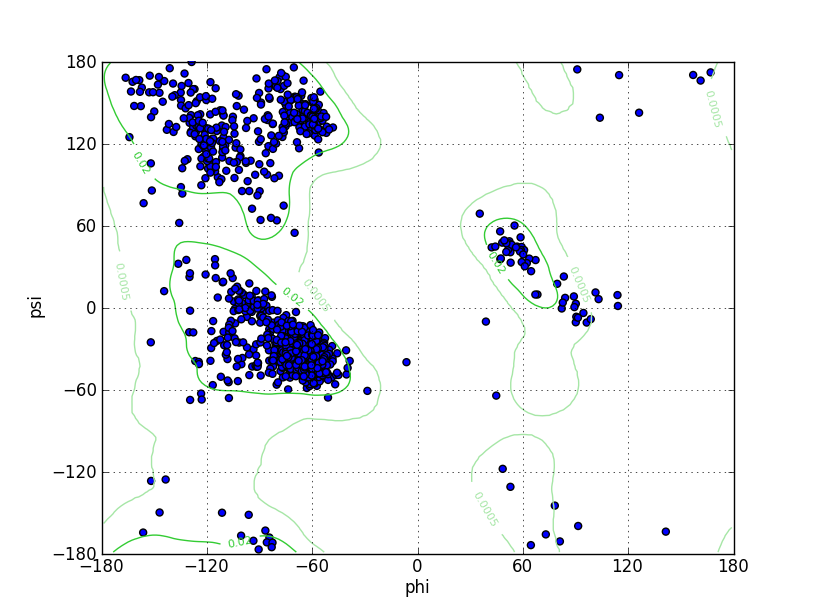
Ramz Z Score:-0.69 ±0.2 Residues in Favored Region:1500 Ramachandran favored:98.10% Number of Outliers:5 Ramachandran outliers:0.33% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
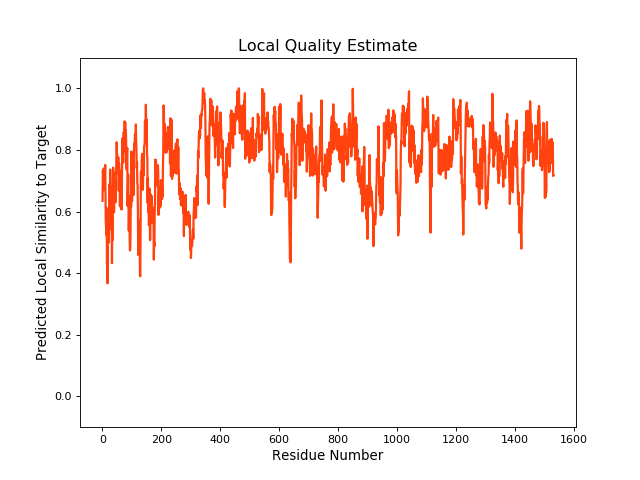
QMEANBrane Score:0.78 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| References | |||||
| 1 | Structural Basis of Substrate Recognition by the Multidrug Resistance Protein MRP1. Cell. 2017 Mar 9;168(6):1075-1085.e9. | ||||
| 2 | ATP Binding Enables Substrate Release from Multidrug Resistance Protein 1. Cell. 2018 Jan 11;172(1-2):81-89.e10. | ||||
| 3 | A macrocyclic peptide inhibitor traps MRP1 in a catalytically incompetent conformation. Proc Natl Acad Sci U S A. 2023;120(11):e2220012120. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.