Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0481 Transporter Info | ||||
| Gene Name | SLC9A1 | ||||
| Transporter Name | Sodium/hydrogen exchanger 1 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Pancretic ductal adenocarcinoma |
13 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC9A1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg08639339) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.58E+00 | Statistic Test | p-value:4.79E-10; Z-score:-1.87E+00 | ||
|
Methylation in Case |
3.81E-01 (Median) | Methylation in Control | 6.03E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC9A1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg07186154) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.56E+00 | Statistic Test | p-value:2.58E-03; Z-score:6.08E-01 | ||
|
Methylation in Case |
1.63E-01 (Median) | Methylation in Control | 1.05E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC9A1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg23346773) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.41E+00 | Statistic Test | p-value:1.08E-04; Z-score:-1.03E+00 | ||
|
Methylation in Case |
1.25E-01 (Median) | Methylation in Control | 1.77E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC9A1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg07966717) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:1.21E-03; Z-score:-5.55E-01 | ||
|
Methylation in Case |
7.71E-02 (Median) | Methylation in Control | 8.63E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC9A1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg17526887) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.23E+00 | Statistic Test | p-value:4.39E-08; Z-score:-1.37E+00 | ||
|
Methylation in Case |
5.09E-01 (Median) | Methylation in Control | 6.26E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC9A1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg01561807) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:4.39E-05; Z-score:-1.38E+00 | ||
|
Methylation in Case |
7.15E-01 (Median) | Methylation in Control | 8.70E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC9A1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg05603680) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:1.75E-04; Z-score:-8.42E-01 | ||
|
Methylation in Case |
5.74E-01 (Median) | Methylation in Control | 6.52E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC9A1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg05230567) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:8.36E-04; Z-score:-1.04E+00 | ||
|
Methylation in Case |
4.43E-01 (Median) | Methylation in Control | 6.00E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC9A1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg02262187) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.43E+00 | Statistic Test | p-value:1.86E-03; Z-score:-1.08E+00 | ||
|
Methylation in Case |
1.81E-01 (Median) | Methylation in Control | 2.59E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC9A1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg14688430) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:2.82E-03; Z-score:9.48E-01 | ||
|
Methylation in Case |
7.56E-01 (Median) | Methylation in Control | 6.99E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC9A1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg07374465) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:2.34E-02; Z-score:2.91E-01 | ||
|
Methylation in Case |
8.91E-01 (Median) | Methylation in Control | 8.85E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC9A1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg05393733) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:3.28E-02; Z-score:2.65E-01 | ||
|
Methylation in Case |
8.52E-01 (Median) | Methylation in Control | 8.38E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC9A1 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg10374523) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:3.67E-02; Z-score:4.01E-01 | ||
|
Methylation in Case |
3.39E-01 (Median) | Methylation in Control | 3.01E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
16 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC9A1 in breast cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg15076659) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:4.63E-03; Z-score:5.20E-01 | ||
|
Methylation in Case |
5.85E-02 (Median) | Methylation in Control | 4.85E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC9A1 in breast cancer | [ 2 ] | |||
|
Location |
1stExon (cg26515162) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:4.75E-02; Z-score:7.30E-01 | ||
|
Methylation in Case |
8.80E-01 (Median) | Methylation in Control | 8.48E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC9A1 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg07359379) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.62E+00 | Statistic Test | p-value:6.92E-31; Z-score:3.85E+00 | ||
|
Methylation in Case |
6.98E-01 (Median) | Methylation in Control | 4.32E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC9A1 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg24738346) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.98E+00 | Statistic Test | p-value:1.85E-18; Z-score:4.09E+00 | ||
|
Methylation in Case |
4.80E-01 (Median) | Methylation in Control | 2.42E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC9A1 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg00456216) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:1.72E-11; Z-score:1.90E+00 | ||
|
Methylation in Case |
6.49E-01 (Median) | Methylation in Control | 5.70E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC9A1 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg15030789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.25E+00 | Statistic Test | p-value:3.31E-08; Z-score:-1.58E+00 | ||
|
Methylation in Case |
5.73E-01 (Median) | Methylation in Control | 7.15E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC9A1 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg09725874) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.28E+00 | Statistic Test | p-value:6.03E-06; Z-score:1.50E+00 | ||
|
Methylation in Case |
3.47E-01 (Median) | Methylation in Control | 2.72E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC9A1 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg05778820) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:8.30E-04; Z-score:1.23E+00 | ||
|
Methylation in Case |
6.75E-01 (Median) | Methylation in Control | 6.07E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC9A1 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg04912273) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:1.47E-03; Z-score:1.16E+00 | ||
|
Methylation in Case |
7.57E-01 (Median) | Methylation in Control | 6.91E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC9A1 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg25130381) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:1.89E-03; Z-score:3.18E-01 | ||
|
Methylation in Case |
7.34E-01 (Median) | Methylation in Control | 7.07E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC9A1 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg13300580) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:6.54E-03; Z-score:5.09E-01 | ||
|
Methylation in Case |
6.87E-01 (Median) | Methylation in Control | 6.63E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC9A1 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg25098478) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:9.05E-03; Z-score:1.23E+00 | ||
|
Methylation in Case |
4.60E-01 (Median) | Methylation in Control | 4.11E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC9A1 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg20816789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.16E-02; Z-score:-8.76E-01 | ||
|
Methylation in Case |
6.22E-01 (Median) | Methylation in Control | 6.58E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC9A1 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg22488568) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:1.24E-02; Z-score:1.48E+00 | ||
|
Methylation in Case |
9.45E-01 (Median) | Methylation in Control | 9.00E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC9A1 in breast cancer | [ 2 ] | |||
|
Location |
3'UTR (cg11673687) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.35E+00 | Statistic Test | p-value:1.58E-14; Z-score:2.20E+00 | ||
|
Methylation in Case |
5.80E-01 (Median) | Methylation in Control | 4.28E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC9A1 in breast cancer | [ 2 ] | |||
|
Location |
3'UTR (cg20942867) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.46E+00 | Statistic Test | p-value:2.93E-05; Z-score:-1.17E+00 | ||
|
Methylation in Case |
1.38E-01 (Median) | Methylation in Control | 2.02E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC9A1 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
TSS1500 (cg11743054) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.28E+00 | Statistic Test | p-value:1.90E-06; Z-score:1.46E+00 | ||
|
Methylation in Case |
2.44E-02 (Median) | Methylation in Control | 1.91E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC9A1 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
TSS1500 (cg17856610) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:7.94E-05; Z-score:9.32E-01 | ||
|
Methylation in Case |
3.02E-02 (Median) | Methylation in Control | 2.69E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC9A1 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
1stExon (cg22481448) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:6.05E-09; Z-score:1.76E+00 | ||
|
Methylation in Case |
9.57E-01 (Median) | Methylation in Control | 8.98E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC9A1 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
1stExon (cg18594482) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:1.05E-04; Z-score:2.92E+00 | ||
|
Methylation in Case |
9.65E-01 (Median) | Methylation in Control | 8.96E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC9A1 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
1stExon (cg26515162) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:9.10E-03; Z-score:1.15E+00 | ||
|
Methylation in Case |
9.64E-01 (Median) | Methylation in Control | 9.49E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colon cancer |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC9A1 in colon adenocarcinoma | [ 4 ] | |||
|
Location |
TSS1500 (cg06066676) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.19E+00 | Statistic Test | p-value:2.12E-06; Z-score:-4.94E+00 | ||
|
Methylation in Case |
6.60E-01 (Median) | Methylation in Control | 7.83E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC9A1 in colon adenocarcinoma | [ 4 ] | |||
|
Location |
TSS1500 (cg04974130) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.54E+00 | Statistic Test | p-value:2.80E-05; Z-score:3.41E+00 | ||
|
Methylation in Case |
3.62E-01 (Median) | Methylation in Control | 2.34E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC9A1 in colon adenocarcinoma | [ 4 ] | |||
|
Location |
Body (cg21602614) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.52E+00 | Statistic Test | p-value:1.30E-07; Z-score:-2.28E+00 | ||
|
Methylation in Case |
2.24E-01 (Median) | Methylation in Control | 3.39E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC9A1 in colon adenocarcinoma | [ 4 ] | |||
|
Location |
Body (cg23023466) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:3.19E-05; Z-score:-1.43E+00 | ||
|
Methylation in Case |
1.30E-01 (Median) | Methylation in Control | 1.76E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC9A1 in colon adenocarcinoma | [ 4 ] | |||
|
Location |
Body (cg27005847) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:1.37E-03; Z-score:-1.15E+00 | ||
|
Methylation in Case |
4.81E-01 (Median) | Methylation in Control | 5.47E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
10 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC9A1 in colorectal cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg15076659) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:1.47E-03; Z-score:8.21E-01 | ||
|
Methylation in Case |
8.99E-02 (Median) | Methylation in Control | 7.57E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC9A1 in colorectal cancer | [ 5 ] | |||
|
Location |
1stExon (cg18594482) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:1.37E-02; Z-score:-4.06E-01 | ||
|
Methylation in Case |
9.09E-01 (Median) | Methylation in Control | 9.52E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC9A1 in colorectal cancer | [ 5 ] | |||
|
Location |
Body (cg15030789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:1.01E-10; Z-score:-2.84E+00 | ||
|
Methylation in Case |
7.49E-01 (Median) | Methylation in Control | 8.51E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC9A1 in colorectal cancer | [ 5 ] | |||
|
Location |
Body (cg11244695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:3.72E-06; Z-score:-2.01E+00 | ||
|
Methylation in Case |
9.24E-01 (Median) | Methylation in Control | 9.44E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC9A1 in colorectal cancer | [ 5 ] | |||
|
Location |
Body (cg05778820) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:4.61E-05; Z-score:1.03E+00 | ||
|
Methylation in Case |
8.57E-01 (Median) | Methylation in Control | 7.59E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC9A1 in colorectal cancer | [ 5 ] | |||
|
Location |
Body (cg09725874) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:1.62E-04; Z-score:1.09E+00 | ||
|
Methylation in Case |
4.38E-01 (Median) | Methylation in Control | 3.58E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC9A1 in colorectal cancer | [ 5 ] | |||
|
Location |
Body (cg13300580) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.13E-03; Z-score:-7.22E-01 | ||
|
Methylation in Case |
8.62E-01 (Median) | Methylation in Control | 8.83E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC9A1 in colorectal cancer | [ 5 ] | |||
|
Location |
Body (cg25130381) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:3.87E-03; Z-score:-1.08E+00 | ||
|
Methylation in Case |
8.21E-01 (Median) | Methylation in Control | 8.61E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC9A1 in colorectal cancer | [ 5 ] | |||
|
Location |
Body (cg00456216) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:3.08E-02; Z-score:-6.71E-01 | ||
|
Methylation in Case |
7.29E-01 (Median) | Methylation in Control | 7.57E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
16 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC9A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg20473622) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.38E+00 | Statistic Test | p-value:1.34E-18; Z-score:-3.47E+00 | ||
|
Methylation in Case |
4.90E-01 (Median) | Methylation in Control | 6.74E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC9A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
TSS200 (cg22367705) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.45E+00 | Statistic Test | p-value:3.83E-02; Z-score:2.73E-01 | ||
|
Methylation in Case |
7.33E-02 (Median) | Methylation in Control | 5.04E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC9A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
1stExon (cg18594482) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:7.44E-03; Z-score:-2.90E-01 | ||
|
Methylation in Case |
9.47E-01 (Median) | Methylation in Control | 9.49E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC9A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
1stExon (cg26515162) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:4.72E-02; Z-score:-1.99E-01 | ||
|
Methylation in Case |
9.54E-01 (Median) | Methylation in Control | 9.57E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC9A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg18677812) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.82E+00 | Statistic Test | p-value:3.69E-22; Z-score:-7.04E+00 | ||
|
Methylation in Case |
4.41E-01 (Median) | Methylation in Control | 8.03E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC9A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg18886814) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.21E+00 | Statistic Test | p-value:6.19E-19; Z-score:-2.12E+00 | ||
|
Methylation in Case |
5.48E-01 (Median) | Methylation in Control | 6.64E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC9A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg09725874) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:8.28E-08; Z-score:1.27E+00 | ||
|
Methylation in Case |
4.92E-01 (Median) | Methylation in Control | 4.09E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC9A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg00456216) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:9.31E-06; Z-score:-9.91E-01 | ||
|
Methylation in Case |
6.88E-01 (Median) | Methylation in Control | 7.13E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC9A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg25098478) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:1.15E-05; Z-score:1.27E+00 | ||
|
Methylation in Case |
6.61E-01 (Median) | Methylation in Control | 6.09E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC9A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg22488568) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.19E-04; Z-score:-5.81E-01 | ||
|
Methylation in Case |
9.74E-01 (Median) | Methylation in Control | 9.78E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC9A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg25130381) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:4.97E-04; Z-score:-1.13E+00 | ||
|
Methylation in Case |
7.20E-01 (Median) | Methylation in Control | 7.57E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC9A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg11244695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:4.63E-03; Z-score:-4.42E-01 | ||
|
Methylation in Case |
8.53E-01 (Median) | Methylation in Control | 8.77E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC9A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg24738346) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.18E+00 | Statistic Test | p-value:5.49E-03; Z-score:1.11E+00 | ||
|
Methylation in Case |
5.51E-01 (Median) | Methylation in Control | 4.68E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC9A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg17820878) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:8.28E-03; Z-score:-7.43E-01 | ||
|
Methylation in Case |
9.45E-01 (Median) | Methylation in Control | 9.57E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC9A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg13300580) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:4.43E-02; Z-score:5.45E-01 | ||
|
Methylation in Case |
6.49E-01 (Median) | Methylation in Control | 6.31E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC9A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
3'UTR (cg11673687) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.16E-02; Z-score:-2.69E-01 | ||
|
Methylation in Case |
7.62E-01 (Median) | Methylation in Control | 7.72E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
12 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC9A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg11743054) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:1.53E-03; Z-score:-5.09E-01 | ||
|
Methylation in Case |
6.92E-02 (Median) | Methylation in Control | 7.57E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC9A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg26151283) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:4.16E-02; Z-score:2.20E-01 | ||
|
Methylation in Case |
7.55E-02 (Median) | Methylation in Control | 7.39E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC9A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
Body (cg20816789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:1.95E-13; Z-score:-2.05E+00 | ||
|
Methylation in Case |
7.48E-01 (Median) | Methylation in Control | 8.28E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC9A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
Body (cg07359379) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.57E+00 | Statistic Test | p-value:3.75E-10; Z-score:-1.84E+00 | ||
|
Methylation in Case |
3.21E-01 (Median) | Methylation in Control | 5.02E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC9A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
Body (cg22488568) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:4.86E-05; Z-score:6.60E-01 | ||
|
Methylation in Case |
9.40E-01 (Median) | Methylation in Control | 9.32E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC9A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
Body (cg24738346) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:7.18E-05; Z-score:1.27E+00 | ||
|
Methylation in Case |
7.26E-01 (Median) | Methylation in Control | 6.58E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC9A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
Body (cg11244695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.87E-03; Z-score:-7.31E-01 | ||
|
Methylation in Case |
9.44E-01 (Median) | Methylation in Control | 9.51E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC9A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
Body (cg15030789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:4.36E-03; Z-score:-8.70E-01 | ||
|
Methylation in Case |
7.90E-01 (Median) | Methylation in Control | 8.33E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC9A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
Body (cg25130381) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:1.12E-02; Z-score:1.08E+00 | ||
|
Methylation in Case |
7.44E-01 (Median) | Methylation in Control | 7.10E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC9A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
Body (cg25098478) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:4.30E-02; Z-score:7.98E-01 | ||
|
Methylation in Case |
5.45E-01 (Median) | Methylation in Control | 5.12E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC9A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
3'UTR (cg11673687) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.48E+00 | Statistic Test | p-value:2.61E-18; Z-score:-4.56E+00 | ||
|
Methylation in Case |
4.84E-01 (Median) | Methylation in Control | 7.18E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC9A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
3'UTR (cg20942867) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.40E+00 | Statistic Test | p-value:6.42E-14; Z-score:-1.75E+00 | ||
|
Methylation in Case |
1.42E-01 (Median) | Methylation in Control | 2.00E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC9A1 in prostate cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg05996789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:9.37E-03; Z-score:-7.57E+00 | ||
|
Methylation in Case |
7.73E-01 (Median) | Methylation in Control | 8.95E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC9A1 in prostate cancer | [ 8 ] | |||
|
Location |
Body (cg25970471) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:5.20E-03; Z-score:3.20E+00 | ||
|
Methylation in Case |
8.57E-01 (Median) | Methylation in Control | 7.71E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC9A1 in prostate cancer | [ 8 ] | |||
|
Location |
Body (cg17434453) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:1.87E-02; Z-score:2.16E+00 | ||
|
Methylation in Case |
9.28E-01 (Median) | Methylation in Control | 9.00E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC9A1 in prostate cancer | [ 8 ] | |||
|
Location |
Body (cg12228245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:4.12E+00 | Statistic Test | p-value:4.73E-02; Z-score:1.97E+01 | ||
|
Methylation in Case |
1.97E-01 (Median) | Methylation in Control | 4.77E-02 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC9A1 in prostate cancer | [ 8 ] | |||
|
Location |
3'UTR (cg16785291) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.30E+00 | Statistic Test | p-value:1.94E-03; Z-score:3.38E+00 | ||
|
Methylation in Case |
8.17E-01 (Median) | Methylation in Control | 6.28E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC9A1 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
TSS200 (cg22367705) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.30E+00 | Statistic Test | p-value:1.71E-03; Z-score:1.76E+00 | ||
|
Methylation in Case |
2.02E-01 (Median) | Methylation in Control | 1.55E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC9A1 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg24738346) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.39E+00 | Statistic Test | p-value:2.08E-04; Z-score:3.75E+00 | ||
|
Methylation in Case |
6.57E-01 (Median) | Methylation in Control | 4.74E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC9A1 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg25130381) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.16E+00 | Statistic Test | p-value:3.07E-03; Z-score:1.75E+00 | ||
|
Methylation in Case |
7.26E-01 (Median) | Methylation in Control | 6.28E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC9A1 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg13300580) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:3.50E-03; Z-score:1.81E+00 | ||
|
Methylation in Case |
7.06E-01 (Median) | Methylation in Control | 6.25E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC9A1 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
3'UTR (cg20942867) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.86E+00 | Statistic Test | p-value:4.12E-04; Z-score:-2.02E+00 | ||
|
Methylation in Case |
1.58E-01 (Median) | Methylation in Control | 2.93E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
16 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC9A1 in bladder cancer | [ 10 ] | |||
|
Location |
1stExon (cg22481448) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-4.14E+00 | Statistic Test | p-value:2.18E-16; Z-score:-2.37E+01 | ||
|
Methylation in Case |
2.17E-01 (Median) | Methylation in Control | 8.96E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC9A1 in bladder cancer | [ 10 ] | |||
|
Location |
1stExon (cg18594482) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.87E+00 | Statistic Test | p-value:5.30E-15; Z-score:-1.47E+01 | ||
|
Methylation in Case |
4.63E-01 (Median) | Methylation in Control | 8.64E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC9A1 in bladder cancer | [ 10 ] | |||
|
Location |
1stExon (cg26515162) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.58E+00 | Statistic Test | p-value:2.95E-14; Z-score:-2.34E+01 | ||
|
Methylation in Case |
3.51E-01 (Median) | Methylation in Control | 9.06E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC9A1 in bladder cancer | [ 10 ] | |||
|
Location |
Body (cg00456216) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.27E+00 | Statistic Test | p-value:2.11E-07; Z-score:6.32E+00 | ||
|
Methylation in Case |
6.34E-01 (Median) | Methylation in Control | 4.98E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC9A1 in bladder cancer | [ 10 ] | |||
|
Location |
Body (cg25130381) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:4.29E-07; Z-score:6.82E+00 | ||
|
Methylation in Case |
8.13E-01 (Median) | Methylation in Control | 7.08E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC9A1 in bladder cancer | [ 10 ] | |||
|
Location |
Body (cg07359379) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.53E+00 | Statistic Test | p-value:3.32E-06; Z-score:5.31E+00 | ||
|
Methylation in Case |
7.68E-01 (Median) | Methylation in Control | 5.03E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC9A1 in bladder cancer | [ 10 ] | |||
|
Location |
Body (cg22488568) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:1.38E-05; Z-score:5.31E+00 | ||
|
Methylation in Case |
9.70E-01 (Median) | Methylation in Control | 8.99E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC9A1 in bladder cancer | [ 10 ] | |||
|
Location |
Body (cg26313337) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.73E+00 | Statistic Test | p-value:4.21E-05; Z-score:-5.06E+00 | ||
|
Methylation in Case |
3.33E-01 (Median) | Methylation in Control | 5.77E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC9A1 in bladder cancer | [ 10 ] | |||
|
Location |
Body (cg11244695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:7.57E-05; Z-score:-3.88E+00 | ||
|
Methylation in Case |
8.45E-01 (Median) | Methylation in Control | 8.91E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC9A1 in bladder cancer | [ 10 ] | |||
|
Location |
Body (cg25098478) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:1.16E-04; Z-score:-5.24E+00 | ||
|
Methylation in Case |
3.36E-01 (Median) | Methylation in Control | 4.54E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC9A1 in bladder cancer | [ 10 ] | |||
|
Location |
Body (cg13300580) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:6.72E-04; Z-score:3.39E+00 | ||
|
Methylation in Case |
7.38E-01 (Median) | Methylation in Control | 6.78E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC9A1 in bladder cancer | [ 10 ] | |||
|
Location |
Body (cg04912273) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:1.14E-03; Z-score:2.49E+00 | ||
|
Methylation in Case |
8.13E-01 (Median) | Methylation in Control | 7.44E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC9A1 in bladder cancer | [ 10 ] | |||
|
Location |
Body (cg09725874) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.15E+00 | Statistic Test | p-value:2.08E-03; Z-score:-2.71E+00 | ||
|
Methylation in Case |
1.48E-01 (Median) | Methylation in Control | 3.19E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC9A1 in bladder cancer | [ 10 ] | |||
|
Location |
Body (cg24738346) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.00E+00 | Statistic Test | p-value:5.63E-03; Z-score:-2.42E+00 | ||
|
Methylation in Case |
2.02E-01 (Median) | Methylation in Control | 4.03E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC9A1 in bladder cancer | [ 10 ] | |||
|
Location |
Body (cg05778820) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.40E+00 | Statistic Test | p-value:1.56E-02; Z-score:2.09E+00 | ||
|
Methylation in Case |
6.20E-01 (Median) | Methylation in Control | 4.43E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC9A1 in bladder cancer | [ 10 ] | |||
|
Location |
3'UTR (cg11673687) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.56E+00 | Statistic Test | p-value:3.73E-05; Z-score:4.28E+00 | ||
|
Methylation in Case |
6.84E-01 (Median) | Methylation in Control | 4.39E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC9A1 in panic disorder | [ 11 ] | |||
|
Location |
1stExon (cg22481448) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:4.68E-02; Z-score:5.50E-01 | ||
|
Methylation in Case |
4.71E+00 (Median) | Methylation in Control | 4.55E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC9A1 in panic disorder | [ 11 ] | |||
|
Location |
Body (cg20816789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:3.62E-03; Z-score:7.74E-01 | ||
|
Methylation in Case |
1.58E+00 (Median) | Methylation in Control | 1.37E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC9A1 in panic disorder | [ 11 ] | |||
|
Location |
Body (cg04912273) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:3.63E-03; Z-score:-4.54E-01 | ||
|
Methylation in Case |
2.91E+00 (Median) | Methylation in Control | 3.06E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC9A1 in panic disorder | [ 11 ] | |||
|
Location |
Body (cg05778820) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:4.08E-02; Z-score:-2.01E-01 | ||
|
Methylation in Case |
3.23E+00 (Median) | Methylation in Control | 3.29E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC9A1 in panic disorder | [ 11 ] | |||
|
Location |
Body (cg11244695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:4.12E-02; Z-score:3.56E-01 | ||
|
Methylation in Case |
3.13E+00 (Median) | Methylation in Control | 2.95E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC9A1 in panic disorder | [ 11 ] | |||
|
Location |
3'UTR (cg20942867) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-9.56E-01 | Statistic Test | p-value:2.71E-02; Z-score:-3.58E-01 | ||
|
Methylation in Case |
-4.51E+00 (Median) | Methylation in Control | -4.31E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
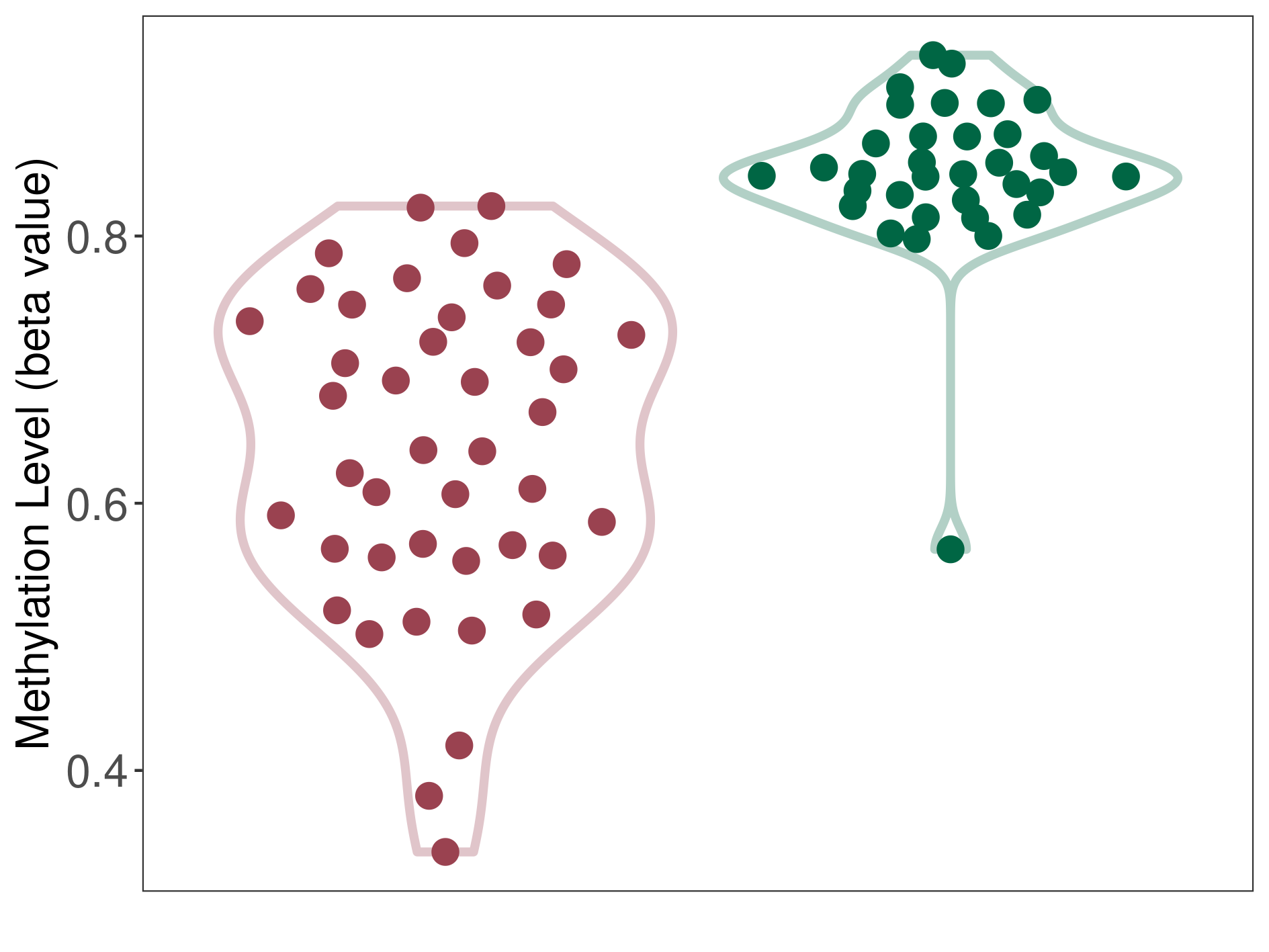
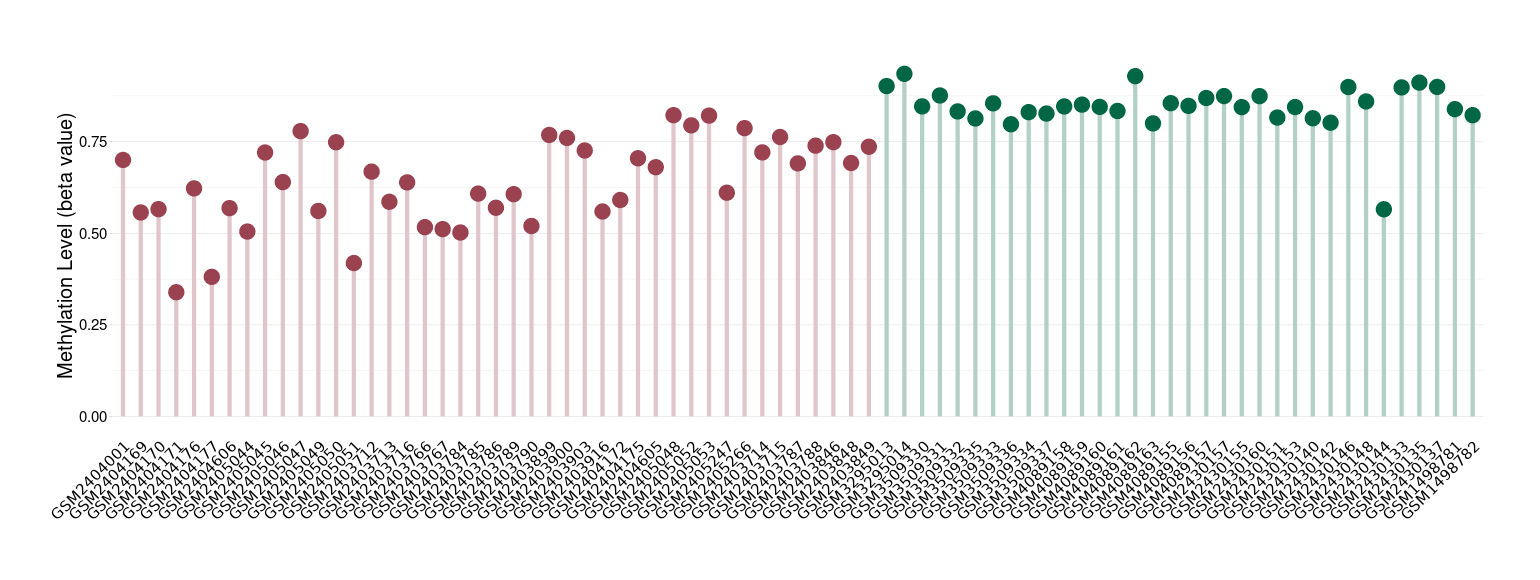
Craniopharyngioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC9A1 in craniopharyngioma than that in healthy individual | ||||
Studied Phenotype |
Craniopharyngioma [ICD-11:2F9A] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.68E-14; Fold-change:-0.206638728; Z-score:-3.3739486 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
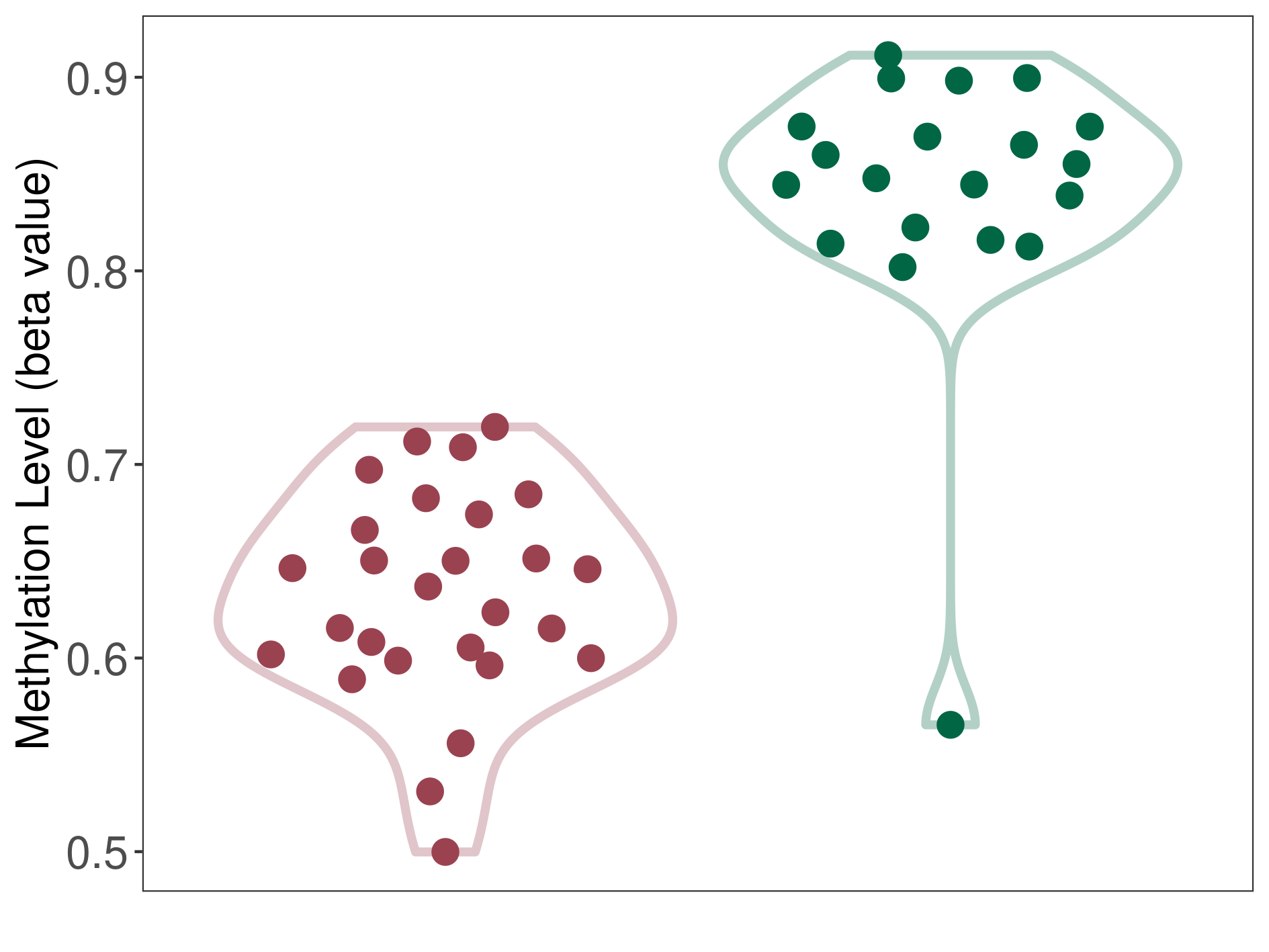
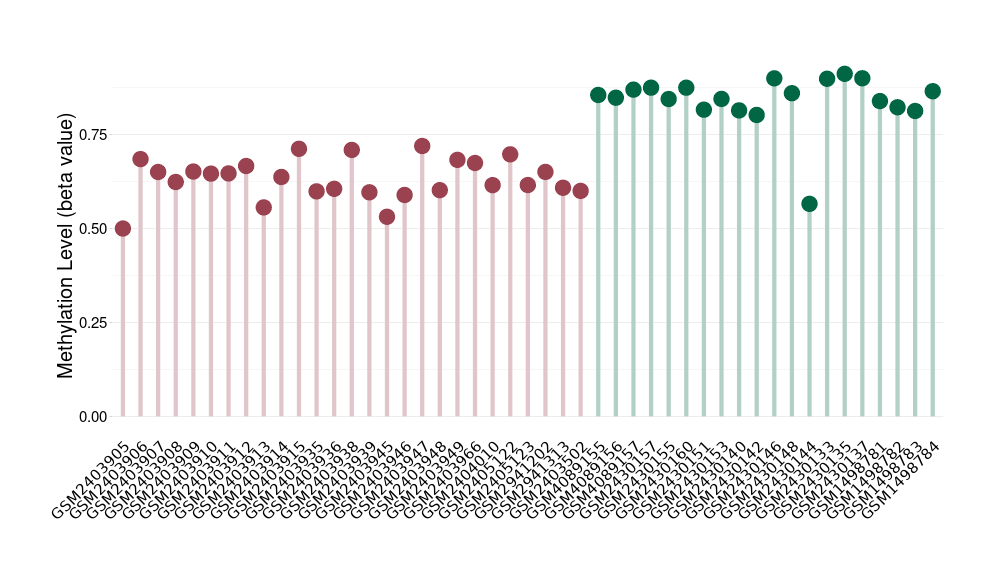
Hemangioblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC9A1 in hemangioblastoma than that in healthy individual | ||||
Studied Phenotype |
Hemangioblastoma [ICD-11:2F7C] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.50E-12; Fold-change:-0.214569817; Z-score:-2.968539067 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
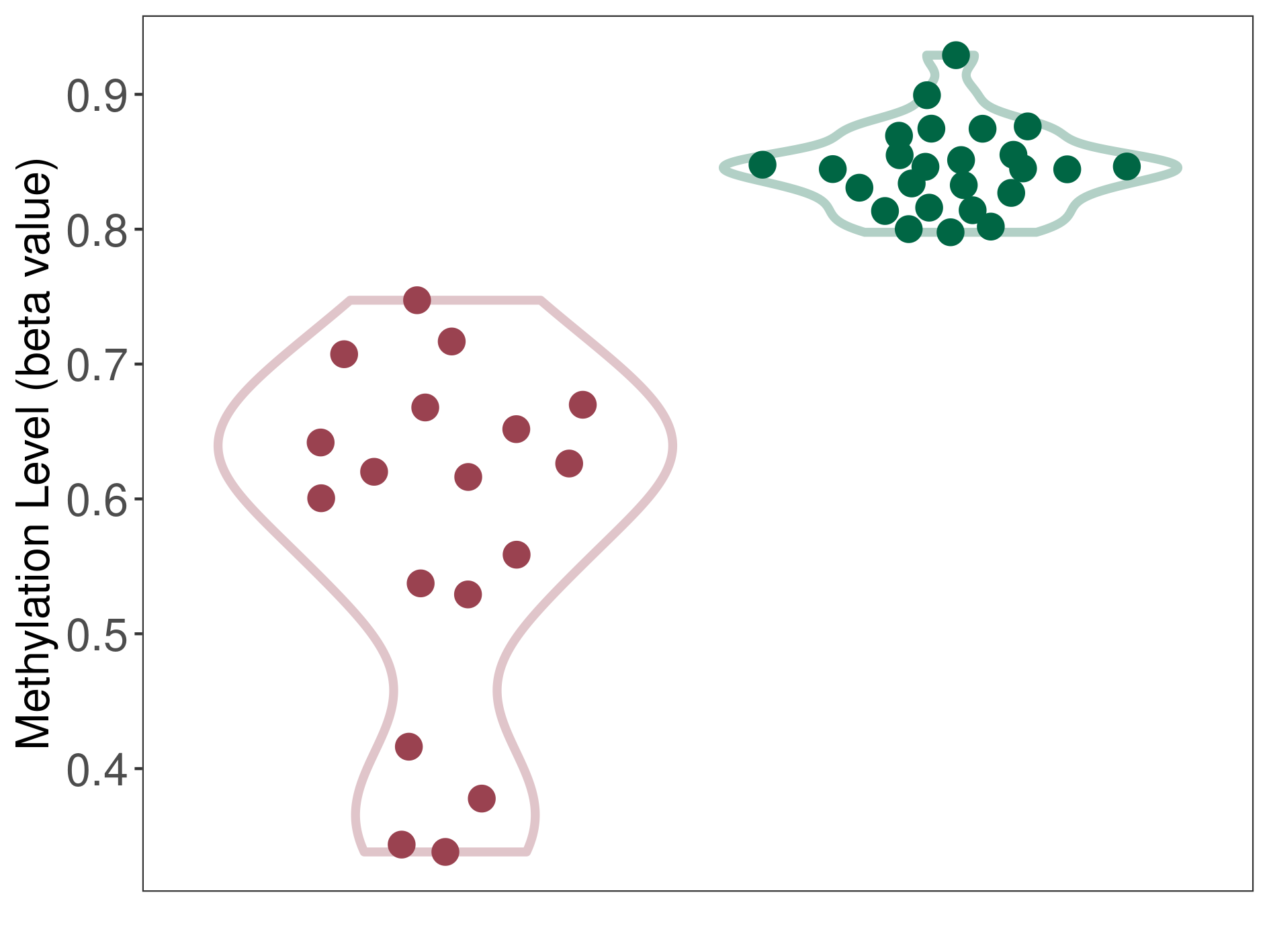
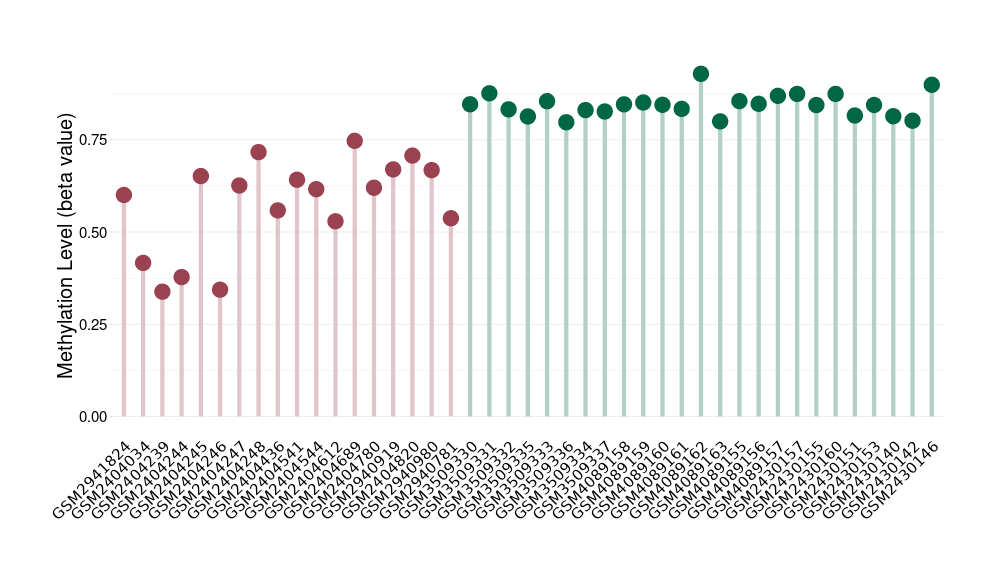
Melanocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC9A1 in melanocytoma than that in healthy individual | ||||
Studied Phenotype |
Melanocytoma [ICD-11:2F36.2] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:5.78E-08; Fold-change:-0.226874282; Z-score:-7.283139691 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
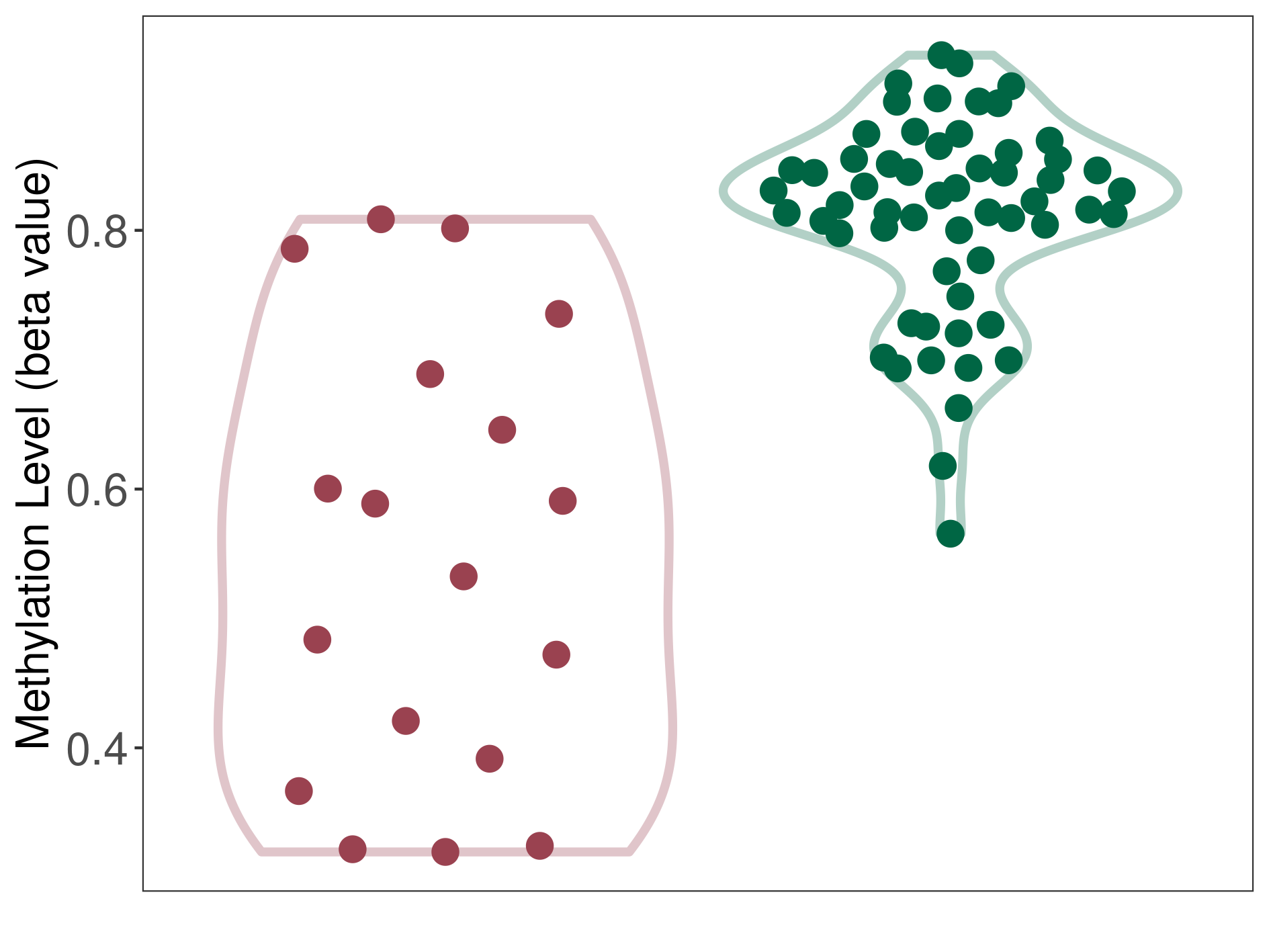
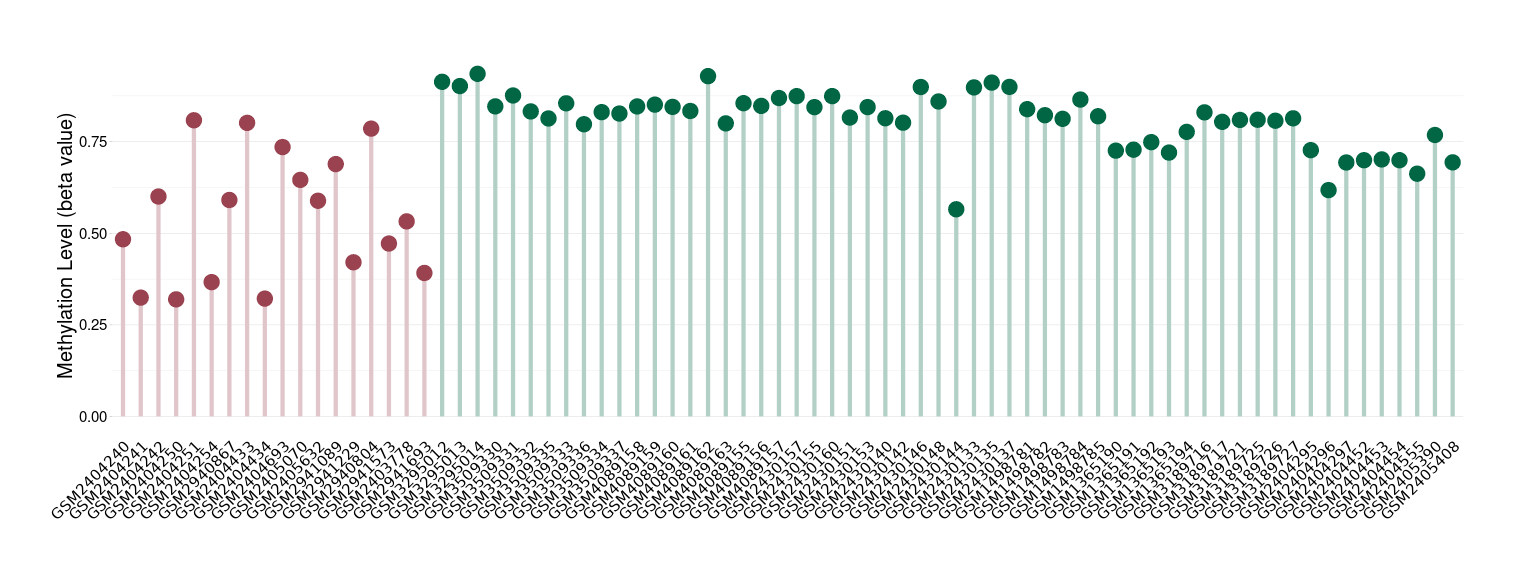
Melanoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC9A1 in melanoma than that in healthy individual | ||||
Studied Phenotype |
Melanoma [ICD-11:2C30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:4.56E-06; Fold-change:-0.264019576; Z-score:-3.423107077 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Peripheral neuroectodermal tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC9A1 in peripheral neuroectodermal tumour than that in healthy individual | ||||
Studied Phenotype |
Peripheral neuroectodermal tumour [ICD-11:2B52] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:3.39E-08; Fold-change:-0.279275043; Z-score:-4.348504017 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
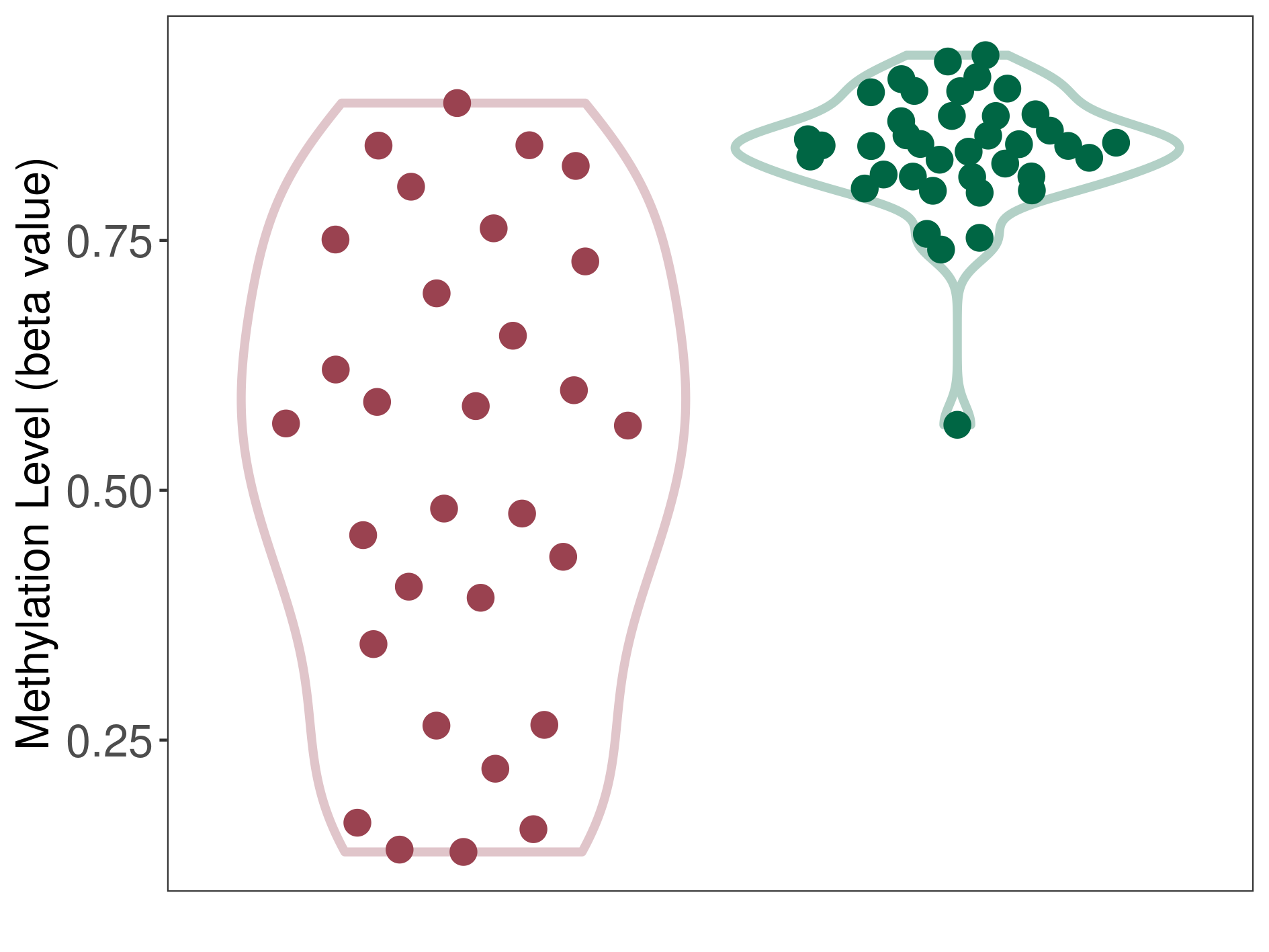
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Posterior fossa ependymoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC9A1 in posterior fossa ependymoma than that in healthy individual | ||||
Studied Phenotype |
Posterior fossa ependymoma [ICD-11:2D50.2] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:4.12E-64; Fold-change:-0.206737652; Z-score:-2.399968945 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
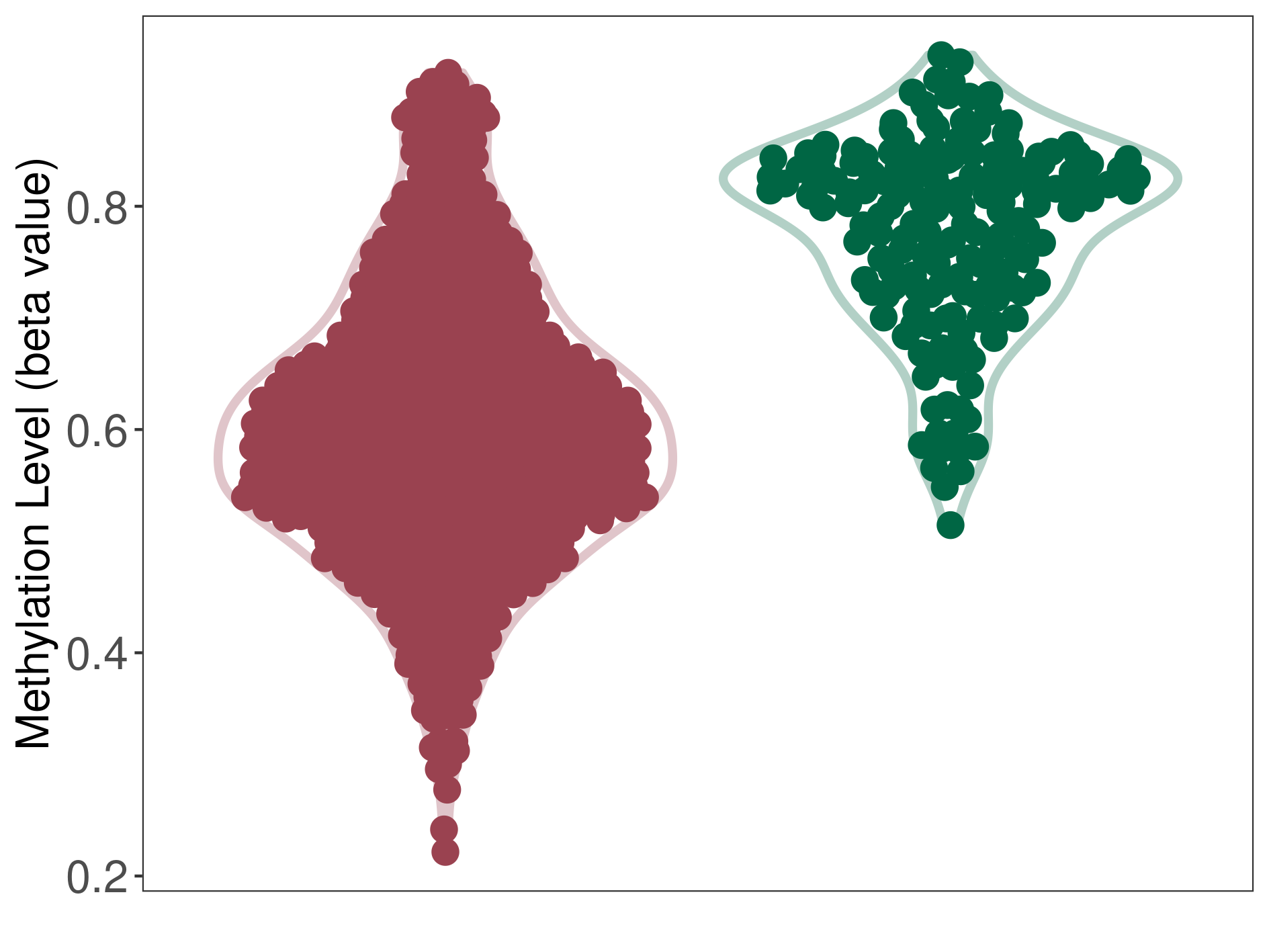
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Brain neuroblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC9A1 in brain neuroblastoma than that in healthy individual | ||||
Studied Phenotype |
Brain neuroblastoma [ICD-11:2A00.11] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.17E-15; Fold-change:-0.326495897; Z-score:-5.033273764 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
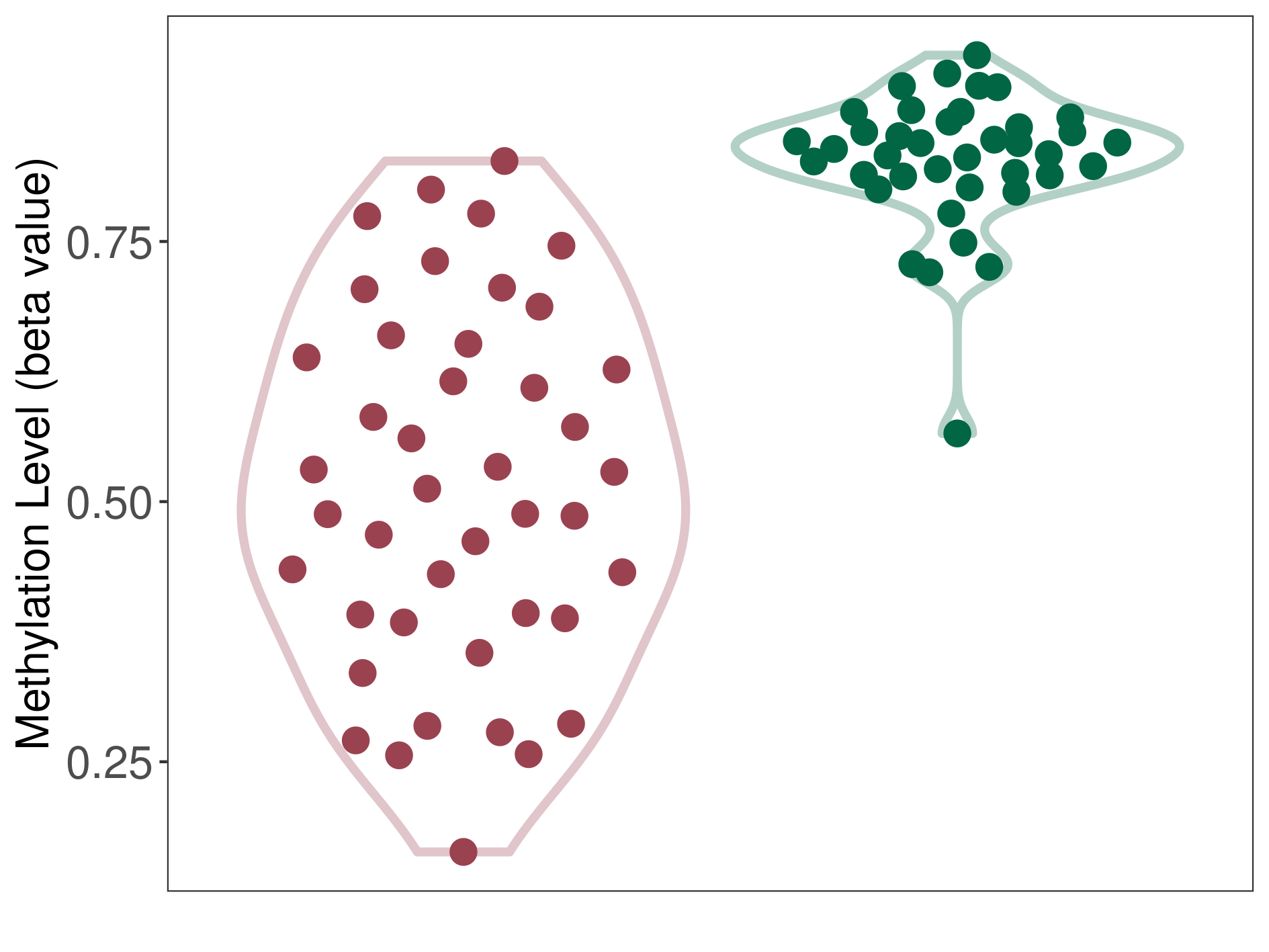
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Chordoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC9A1 in chordoma than that in healthy individual | ||||
Studied Phenotype |
Chordoma [ICD-11:5A61.0] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.53E-07; Fold-change:-0.474358367; Z-score:-5.50334359 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
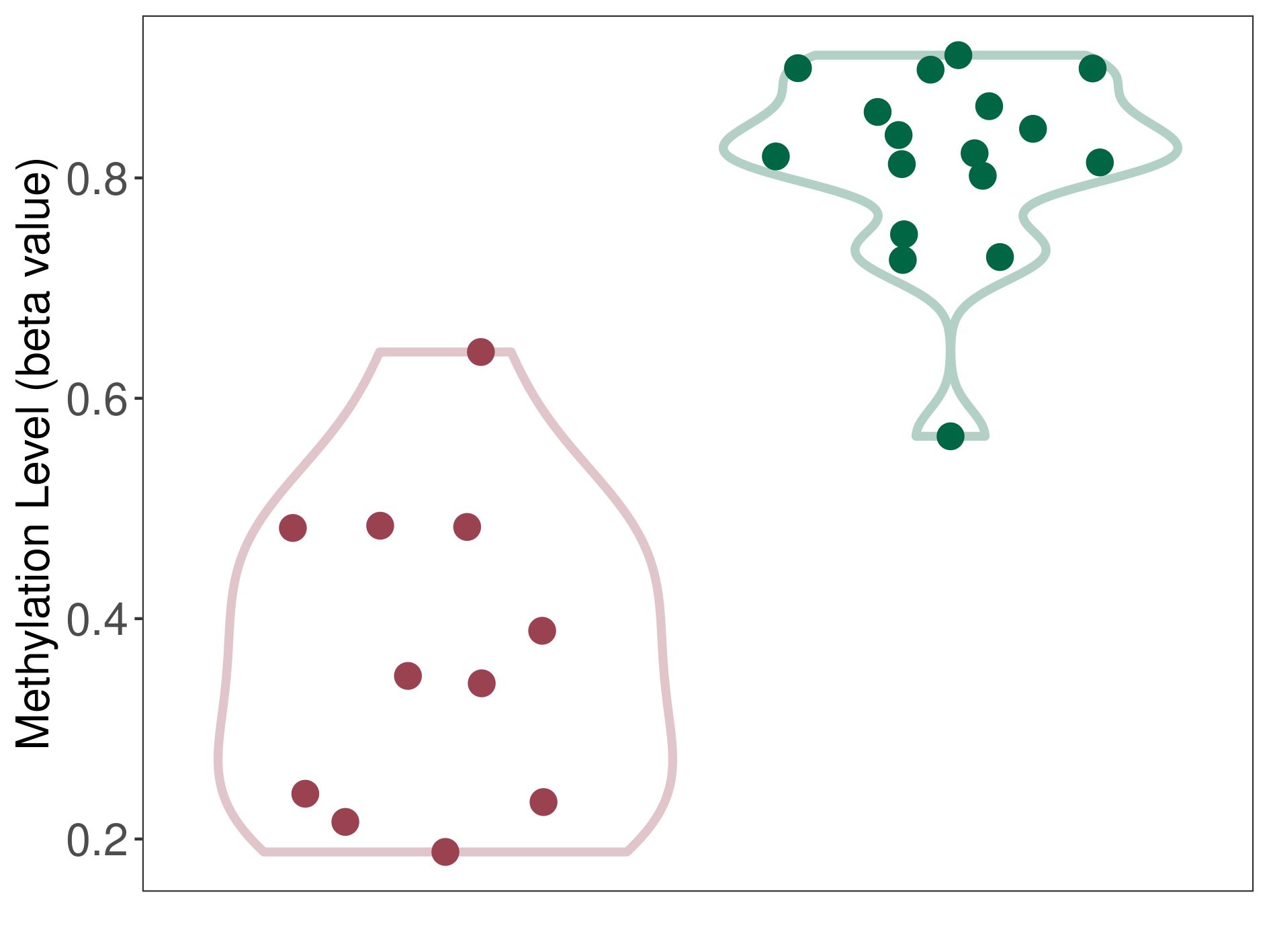
|
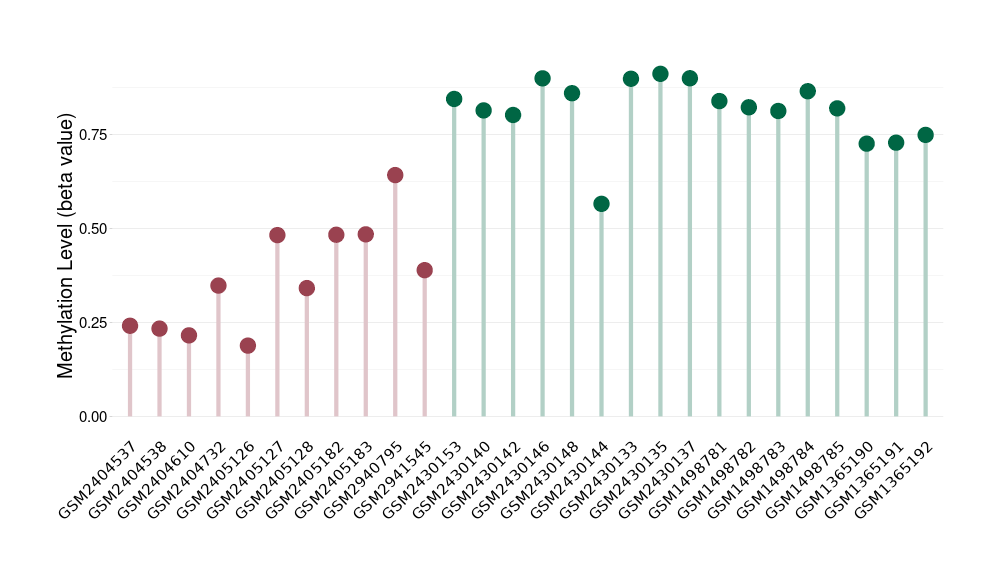 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
microRNA |
|||||
|
Unclear Phenotype |
29 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
miR-106b directly targets SLC9A1 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | CLASH | ||
|
miRNA Stemloop ID |
miR-106b | miRNA Mature ID | miR-106b-5p | ||
|
miRNA Sequence |
UAAAGUGCUGACAGUGCAGAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon2 |
miR-122 directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-122 | miRNA Mature ID | miR-122-5p | ||
|
miRNA Sequence |
UGGAGUGUGACAAUGGUGUUUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon3 |
miR-147a directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-147a | miRNA Mature ID | miR-147a | ||
|
miRNA Sequence |
GUGUGUGGAAAUGCUUCUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon4 |
miR-15a directly targets SLC9A1 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-15a | miRNA Mature ID | miR-15a-5p | ||
|
miRNA Sequence |
UAGCAGCACAUAAUGGUUUGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon5 |
miR-15b directly targets SLC9A1 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-15b | miRNA Mature ID | miR-15b-5p | ||
|
miRNA Sequence |
UAGCAGCACAUCAUGGUUUACA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon6 |
miR-16 directly targets SLC9A1 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-16 | miRNA Mature ID | miR-16-5p | ||
|
miRNA Sequence |
UAGCAGCACGUAAAUAUUGGCG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon7 |
miR-195 directly targets SLC9A1 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-195 | miRNA Mature ID | miR-195-5p | ||
|
miRNA Sequence |
UAGCAGCACAGAAAUAUUGGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon8 |
miR-19a directly targets SLC9A1 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-19a | miRNA Mature ID | miR-19a-3p | ||
|
miRNA Sequence |
UGUGCAAAUCUAUGCAAAACUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon9 |
miR-19b directly targets SLC9A1 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-19b | miRNA Mature ID | miR-19b-3p | ||
|
miRNA Sequence |
UGUGCAAAUCCAUGCAAAACUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon10 |
miR-32 directly targets SLC9A1 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-32 | miRNA Mature ID | miR-32-5p | ||
|
miRNA Sequence |
UAUUGCACAUUACUAAGUUGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon11 |
miR-331 directly targets SLC9A1 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | CLASH | ||
|
miRNA Stemloop ID |
miR-331 | miRNA Mature ID | miR-331-3p | ||
|
miRNA Sequence |
GCCCCUGGGCCUAUCCUAGAA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon12 |
miR-3911 directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3911 | miRNA Mature ID | miR-3911 | ||
|
miRNA Sequence |
UGUGUGGAUCCUGGAGGAGGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon13 |
miR-4265 directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4265 | miRNA Mature ID | miR-4265 | ||
|
miRNA Sequence |
CUGUGGGCUCAGCUCUGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon14 |
miR-4271 directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4271 | miRNA Mature ID | miR-4271 | ||
|
miRNA Sequence |
GGGGGAAGAAAAGGUGGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon15 |
miR-4296 directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4296 | miRNA Mature ID | miR-4296 | ||
|
miRNA Sequence |
AUGUGGGCUCAGGCUCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon16 |
miR-4322 directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4322 | miRNA Mature ID | miR-4322 | ||
|
miRNA Sequence |
CUGUGGGCUCAGCGCGUGGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon17 |
miR-4433b directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4433b | miRNA Mature ID | miR-4433b-3p | ||
|
miRNA Sequence |
CAGGAGUGGGGGGUGGGACGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon18 |
miR-4533 directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4533 | miRNA Mature ID | miR-4533 | ||
|
miRNA Sequence |
UGGAAGGAGGUUGCCGGACGCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon19 |
miR-4725 directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4725 | miRNA Mature ID | miR-4725-3p | ||
|
miRNA Sequence |
UGGGGAAGGCGUCAGUGUCGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon20 |
miR-4747 directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4747 | miRNA Mature ID | miR-4747-5p | ||
|
miRNA Sequence |
AGGGAAGGAGGCUUGGUCUUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon21 |
miR-5196 directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-5196 | miRNA Mature ID | miR-5196-5p | ||
|
miRNA Sequence |
AGGGAAGGGGACGAGGGUUGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon22 |
miR-574 directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-574 | miRNA Mature ID | miR-574-5p | ||
|
miRNA Sequence |
UGAGUGUGUGUGUGUGAGUGUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon23 |
miR-644a directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-644a | miRNA Mature ID | miR-644a | ||
|
miRNA Sequence |
AGUGUGGCUUUCUUAGAGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon24 |
miR-6780b directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6780b | miRNA Mature ID | miR-6780b-5p | ||
|
miRNA Sequence |
UGGGGAAGGCUUGGCAGGGAAGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon25 |
miR-6783 directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6783 | miRNA Mature ID | miR-6783-5p | ||
|
miRNA Sequence |
UAGGGGAAAAGUCCUGAUCCGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon26 |
miR-6867 directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6867 | miRNA Mature ID | miR-6867-5p | ||
|
miRNA Sequence |
UGUGUGUGUAGAGGAAGAAGGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon27 |
miR-7845 directly targets SLC9A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-7845 | miRNA Mature ID | miR-7845-5p | ||
|
miRNA Sequence |
AAGGGACAGGGAGGGUCGUGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon28 |
miR-92a directly targets SLC9A1 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-92a | miRNA Mature ID | miR-92a-3p | ||
|
miRNA Sequence |
UAUUGCACUUGUCCCGGCCUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon29 |
miR-92b directly targets SLC9A1 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-92b | miRNA Mature ID | miR-92b-3p | ||
|
miRNA Sequence |
UAUUGCACUCGUCCCGGCCUCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
