Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0080 Transporter Info | ||||
| Gene Name | SLC11A1 | ||||
| Transporter Name | Natural resistance-associated macrophage protein 1 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Colon cancer |
8 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC11A1 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg26870567) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.17E+00 | Statistic Test | p-value:5.48E-06; Z-score:-2.90E+00 | ||
|
Methylation in Case |
6.51E-01 (Median) | Methylation in Control | 7.62E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC11A1 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg16857852) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.33E+00 | Statistic Test | p-value:1.14E-05; Z-score:-1.67E+00 | ||
|
Methylation in Case |
3.83E-01 (Median) | Methylation in Control | 5.09E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC11A1 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg14788003) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.37E+00 | Statistic Test | p-value:1.04E-06; Z-score:-5.42E+00 | ||
|
Methylation in Case |
5.36E-01 (Median) | Methylation in Control | 7.34E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC11A1 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg14260073) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:9.68E-06; Z-score:8.85E-01 | ||
|
Methylation in Case |
5.54E-01 (Median) | Methylation in Control | 4.93E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC11A1 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg19115882) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.19E+00 | Statistic Test | p-value:5.18E-05; Z-score:-2.10E+00 | ||
|
Methylation in Case |
5.79E-01 (Median) | Methylation in Control | 6.90E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC11A1 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg08490220) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.25E+00 | Statistic Test | p-value:8.79E-05; Z-score:-3.58E+00 | ||
|
Methylation in Case |
5.40E-01 (Median) | Methylation in Control | 6.73E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC11A1 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg23516560) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:9.75E-04; Z-score:-1.62E+00 | ||
|
Methylation in Case |
6.72E-01 (Median) | Methylation in Control | 7.46E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC11A1 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg11244695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:3.36E-03; Z-score:-9.18E-01 | ||
|
Methylation in Case |
8.61E-01 (Median) | Methylation in Control | 8.77E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Pancretic ductal adenocarcinoma |
15 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC11A1 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
5'UTR (cg01289541) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.59E+00 | Statistic Test | p-value:7.47E-07; Z-score:-1.55E+00 | ||
|
Methylation in Case |
3.94E-01 (Median) | Methylation in Control | 6.27E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC11A1 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg02313331) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.53E+00 | Statistic Test | p-value:1.03E-06; Z-score:-1.29E+00 | ||
|
Methylation in Case |
2.41E-01 (Median) | Methylation in Control | 3.68E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC11A1 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg07583503) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:2.89E-04; Z-score:3.27E-01 | ||
|
Methylation in Case |
2.55E-01 (Median) | Methylation in Control | 2.34E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC11A1 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg11455040) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:2.64E-03; Z-score:-3.80E-01 | ||
|
Methylation in Case |
7.51E-02 (Median) | Methylation in Control | 8.64E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC11A1 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg19616230) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.31E+00 | Statistic Test | p-value:3.57E-11; Z-score:-2.04E+00 | ||
|
Methylation in Case |
2.10E-01 (Median) | Methylation in Control | 2.75E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC11A1 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg02615599) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.33E+00 | Statistic Test | p-value:1.32E-07; Z-score:-1.53E+00 | ||
|
Methylation in Case |
1.48E-01 (Median) | Methylation in Control | 1.97E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC11A1 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg10881110) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:1.96E-03; Z-score:6.00E-01 | ||
|
Methylation in Case |
5.95E-01 (Median) | Methylation in Control | 5.43E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC11A1 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
1stExon (cg10671668) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:3.72E+00 | Statistic Test | p-value:2.65E-13; Z-score:2.70E+00 | ||
|
Methylation in Case |
4.88E-01 (Median) | Methylation in Control | 1.31E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC11A1 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg04106903) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:4.01E-05; Z-score:-1.17E+00 | ||
|
Methylation in Case |
1.66E-01 (Median) | Methylation in Control | 2.02E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC11A1 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg13224075) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:1.53E-04; Z-score:9.23E-01 | ||
|
Methylation in Case |
9.36E-01 (Median) | Methylation in Control | 9.02E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC11A1 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg17172593) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:3.27E-03; Z-score:-7.74E-01 | ||
|
Methylation in Case |
4.86E-01 (Median) | Methylation in Control | 5.48E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC11A1 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg05561775) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:9.85E-03; Z-score:5.63E-01 | ||
|
Methylation in Case |
8.77E-01 (Median) | Methylation in Control | 8.62E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC11A1 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg04900941) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.35E-02; Z-score:-3.17E-01 | ||
|
Methylation in Case |
7.62E-01 (Median) | Methylation in Control | 7.78E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC11A1 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg01977413) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.20E+00 | Statistic Test | p-value:1.50E-02; Z-score:-4.97E-01 | ||
|
Methylation in Case |
1.26E-01 (Median) | Methylation in Control | 1.51E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC11A1 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg25423829) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:3.54E-02; Z-score:-6.04E-01 | ||
|
Methylation in Case |
8.43E-01 (Median) | Methylation in Control | 8.57E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
13 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC11A1 in bladder cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg15099037) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.69E+00 | Statistic Test | p-value:7.10E-13; Z-score:-1.50E+01 | ||
|
Methylation in Case |
3.44E-01 (Median) | Methylation in Control | 5.82E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC11A1 in bladder cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg13802355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.54E+00 | Statistic Test | p-value:7.32E-06; Z-score:-4.42E+00 | ||
|
Methylation in Case |
3.14E-01 (Median) | Methylation in Control | 4.82E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC11A1 in bladder cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg09617319) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:5.41E-05; Z-score:-1.19E+01 | ||
|
Methylation in Case |
7.37E-01 (Median) | Methylation in Control | 8.24E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC11A1 in bladder cancer | [ 3 ] | |||
|
Location |
TSS200 (cg18329052) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.94E+00 | Statistic Test | p-value:2.44E-10; Z-score:-1.11E+01 | ||
|
Methylation in Case |
3.41E-01 (Median) | Methylation in Control | 6.61E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC11A1 in bladder cancer | [ 3 ] | |||
|
Location |
TSS200 (cg07719512) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.41E+00 | Statistic Test | p-value:4.65E-10; Z-score:-8.21E+00 | ||
|
Methylation in Case |
3.89E-01 (Median) | Methylation in Control | 5.48E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC11A1 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg16926316) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.06E+00 | Statistic Test | p-value:1.33E-16; Z-score:-2.12E+01 | ||
|
Methylation in Case |
4.26E-01 (Median) | Methylation in Control | 8.78E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC11A1 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg23212579) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.01E+00 | Statistic Test | p-value:3.54E-15; Z-score:-1.12E+02 | ||
|
Methylation in Case |
4.87E-01 (Median) | Methylation in Control | 9.78E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC11A1 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg17130789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.12E+00 | Statistic Test | p-value:5.06E-15; Z-score:-1.59E+01 | ||
|
Methylation in Case |
3.65E-01 (Median) | Methylation in Control | 7.74E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC11A1 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg11378979) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.08E+00 | Statistic Test | p-value:7.76E-10; Z-score:-8.87E+00 | ||
|
Methylation in Case |
3.34E-01 (Median) | Methylation in Control | 6.95E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC11A1 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg04337188) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.28E+00 | Statistic Test | p-value:7.93E-08; Z-score:-1.33E+01 | ||
|
Methylation in Case |
6.13E-01 (Median) | Methylation in Control | 7.87E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC11A1 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg01520853) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:1.02E-06; Z-score:-5.55E+00 | ||
|
Methylation in Case |
7.87E-01 (Median) | Methylation in Control | 8.51E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC11A1 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg17010118) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:1.35E-06; Z-score:-1.19E+01 | ||
|
Methylation in Case |
7.92E-01 (Median) | Methylation in Control | 8.94E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC11A1 in bladder cancer | [ 3 ] | |||
|
Location |
3'UTR (cg24007161) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:7.95E-03; Z-score:-1.32E+00 | ||
|
Methylation in Case |
5.64E-02 (Median) | Methylation in Control | 6.04E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
16 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC11A1 in breast cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg15099037) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.25E+00 | Statistic Test | p-value:1.35E-06; Z-score:-1.45E+00 | ||
|
Methylation in Case |
4.98E-01 (Median) | Methylation in Control | 6.23E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC11A1 in breast cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg13802355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:2.50E-03; Z-score:-6.01E-01 | ||
|
Methylation in Case |
3.66E-01 (Median) | Methylation in Control | 4.20E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC11A1 in breast cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg09617319) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:3.22E-02; Z-score:-5.31E-01 | ||
|
Methylation in Case |
7.88E-01 (Median) | Methylation in Control | 8.18E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC11A1 in breast cancer | [ 4 ] | |||
|
Location |
TSS200 (cg07719512) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:7.85E-04; Z-score:-1.15E+00 | ||
|
Methylation in Case |
5.66E-01 (Median) | Methylation in Control | 6.31E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC11A1 in breast cancer | [ 4 ] | |||
|
Location |
TSS200 (cg18329052) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:1.77E-03; Z-score:-8.31E-01 | ||
|
Methylation in Case |
6.63E-01 (Median) | Methylation in Control | 7.51E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC11A1 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg07015784) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:5.26E-15; Z-score:2.40E+00 | ||
|
Methylation in Case |
9.04E-01 (Median) | Methylation in Control | 7.97E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC11A1 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg11378979) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:1.93E-12; Z-score:-2.72E+00 | ||
|
Methylation in Case |
7.43E-01 (Median) | Methylation in Control | 8.17E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC11A1 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg17130789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:3.33E-06; Z-score:-1.28E+00 | ||
|
Methylation in Case |
7.43E-01 (Median) | Methylation in Control | 7.85E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC11A1 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg04337188) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.17E+00 | Statistic Test | p-value:4.48E-05; Z-score:-1.13E+00 | ||
|
Methylation in Case |
6.68E-01 (Median) | Methylation in Control | 7.81E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC11A1 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg17010118) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:9.77E-05; Z-score:-1.14E+00 | ||
|
Methylation in Case |
8.58E-01 (Median) | Methylation in Control | 8.94E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC11A1 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg00634542) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:1.26E-04; Z-score:-1.08E+00 | ||
|
Methylation in Case |
8.06E-01 (Median) | Methylation in Control | 8.50E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC11A1 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg23212579) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:9.47E-03; Z-score:-5.02E-01 | ||
|
Methylation in Case |
9.64E-01 (Median) | Methylation in Control | 9.79E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC11A1 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg01520853) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.19E-02; Z-score:-6.64E-01 | ||
|
Methylation in Case |
8.42E-01 (Median) | Methylation in Control | 8.59E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC11A1 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg03710889) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:1.34E-02; Z-score:4.36E-01 | ||
|
Methylation in Case |
5.91E-01 (Median) | Methylation in Control | 5.59E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC11A1 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg16926316) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:2.61E-02; Z-score:-5.15E-01 | ||
|
Methylation in Case |
8.38E-01 (Median) | Methylation in Control | 8.84E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC11A1 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg04656051) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:3.49E-02; Z-score:2.82E-01 | ||
|
Methylation in Case |
4.47E-01 (Median) | Methylation in Control | 4.25E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
14 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC11A1 in colorectal cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg15099037) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:2.24E-11; Z-score:-2.30E+00 | ||
|
Methylation in Case |
6.42E-01 (Median) | Methylation in Control | 7.44E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC11A1 in colorectal cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg09617319) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:2.99E-06; Z-score:-1.52E+00 | ||
|
Methylation in Case |
9.02E-01 (Median) | Methylation in Control | 9.22E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC11A1 in colorectal cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg13802355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:1.56E-02; Z-score:7.06E-01 | ||
|
Methylation in Case |
5.74E-01 (Median) | Methylation in Control | 5.15E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC11A1 in colorectal cancer | [ 5 ] | |||
|
Location |
TSS200 (cg07719512) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.19E+00 | Statistic Test | p-value:2.08E-14; Z-score:-3.07E+00 | ||
|
Methylation in Case |
6.51E-01 (Median) | Methylation in Control | 7.78E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC11A1 in colorectal cancer | [ 5 ] | |||
|
Location |
TSS200 (cg18329052) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:4.19E-04; Z-score:-9.35E-01 | ||
|
Methylation in Case |
7.58E-01 (Median) | Methylation in Control | 8.58E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC11A1 in colorectal cancer | [ 5 ] | |||
|
Location |
Body (cg04337188) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:2.46E-10; Z-score:-4.85E+00 | ||
|
Methylation in Case |
8.08E-01 (Median) | Methylation in Control | 8.85E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC11A1 in colorectal cancer | [ 5 ] | |||
|
Location |
Body (cg00634542) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.20E+00 | Statistic Test | p-value:1.82E-08; Z-score:-2.10E+00 | ||
|
Methylation in Case |
6.88E-01 (Median) | Methylation in Control | 8.24E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC11A1 in colorectal cancer | [ 5 ] | |||
|
Location |
Body (cg17130789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:2.98E-08; Z-score:-2.10E+00 | ||
|
Methylation in Case |
8.50E-01 (Median) | Methylation in Control | 8.82E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC11A1 in colorectal cancer | [ 5 ] | |||
|
Location |
Body (cg16926316) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:3.42E-06; Z-score:-1.49E+00 | ||
|
Methylation in Case |
8.79E-01 (Median) | Methylation in Control | 9.07E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC11A1 in colorectal cancer | [ 5 ] | |||
|
Location |
Body (cg23212579) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.19E-03; Z-score:-1.16E+00 | ||
|
Methylation in Case |
9.67E-01 (Median) | Methylation in Control | 9.74E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC11A1 in colorectal cancer | [ 5 ] | |||
|
Location |
Body (cg01520853) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.77E-03; Z-score:-5.17E-01 | ||
|
Methylation in Case |
9.28E-01 (Median) | Methylation in Control | 9.33E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC11A1 in colorectal cancer | [ 5 ] | |||
|
Location |
3'UTR (cg24007161) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.17E+00 | Statistic Test | p-value:3.73E-05; Z-score:-1.05E+00 | ||
|
Methylation in Case |
1.09E-01 (Median) | Methylation in Control | 1.28E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC11A1 in colorectal cancer | [ 5 ] | |||
|
Location |
3'UTR (cg01379701) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.27E+00 | Statistic Test | p-value:1.22E-03; Z-score:-1.00E+00 | ||
|
Methylation in Case |
2.52E-02 (Median) | Methylation in Control | 3.18E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Moderate hypomethylation of SLC11A1 in colorectal cancer than that in healthy individual | ||||
Studied Phenotype |
Colorectal cancer [ICD-11:2B91] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:2.09E-18; Fold-change:-0.204270851; Z-score:-1.734780754 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue adjacent to the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
Hepatocellular carcinoma |
16 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC11A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg15099037) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.23E+00 | Statistic Test | p-value:5.81E-09; Z-score:-2.21E+00 | ||
|
Methylation in Case |
4.46E-01 (Median) | Methylation in Control | 5.50E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC11A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg09617319) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:1.01E-08; Z-score:-1.06E+00 | ||
|
Methylation in Case |
7.21E-01 (Median) | Methylation in Control | 8.33E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC11A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg13802355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.25E-02; Z-score:-1.10E-01 | ||
|
Methylation in Case |
3.85E-01 (Median) | Methylation in Control | 3.98E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC11A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
TSS200 (cg08706670) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:6.20E+00 | Statistic Test | p-value:2.78E-14; Z-score:9.33E+00 | ||
|
Methylation in Case |
3.82E-01 (Median) | Methylation in Control | 6.16E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC11A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
TSS200 (cg26482893) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.97E+00 | Statistic Test | p-value:6.27E-13; Z-score:2.26E+00 | ||
|
Methylation in Case |
6.41E-01 (Median) | Methylation in Control | 3.25E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC11A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
TSS200 (cg07719512) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:6.56E-05; Z-score:-9.56E-01 | ||
|
Methylation in Case |
5.44E-01 (Median) | Methylation in Control | 5.93E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC11A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg00712593) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.62E+00 | Statistic Test | p-value:5.57E-18; Z-score:-6.08E+00 | ||
|
Methylation in Case |
5.05E-01 (Median) | Methylation in Control | 8.17E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC11A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg17780246) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.63E+00 | Statistic Test | p-value:6.46E-16; Z-score:2.86E+00 | ||
|
Methylation in Case |
5.42E-01 (Median) | Methylation in Control | 3.33E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC11A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg19807207) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:1.12E-10; Z-score:-3.47E+00 | ||
|
Methylation in Case |
6.73E-01 (Median) | Methylation in Control | 7.92E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC11A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg04337188) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:1.36E-08; Z-score:-2.26E+00 | ||
|
Methylation in Case |
7.15E-01 (Median) | Methylation in Control | 7.97E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC11A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg11378979) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:8.59E-07; Z-score:-1.43E+00 | ||
|
Methylation in Case |
7.91E-01 (Median) | Methylation in Control | 8.21E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC11A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg17130789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:7.66E-06; Z-score:-1.01E+00 | ||
|
Methylation in Case |
7.91E-01 (Median) | Methylation in Control | 8.13E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC11A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg00634542) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:3.05E-04; Z-score:-6.07E-01 | ||
|
Methylation in Case |
7.51E-01 (Median) | Methylation in Control | 7.95E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC11A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg23212579) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.81E-03; Z-score:-5.76E-01 | ||
|
Methylation in Case |
9.78E-01 (Median) | Methylation in Control | 9.82E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC11A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
Body (cg07015784) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:6.95E-03; Z-score:6.26E-01 | ||
|
Methylation in Case |
9.49E-01 (Median) | Methylation in Control | 9.36E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC11A1 in hepatocellular carcinoma | [ 6 ] | |||
|
Location |
3'UTR (cg22234897) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.68E+00 | Statistic Test | p-value:3.22E-10; Z-score:2.49E+00 | ||
|
Methylation in Case |
1.88E-01 (Median) | Methylation in Control | 6.99E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC11A1 in lung adenocarcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg15099037) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:1.35E-02; Z-score:-1.33E+00 | ||
|
Methylation in Case |
5.78E-01 (Median) | Methylation in Control | 6.44E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC11A1 in lung adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg03291396) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.44E+00 | Statistic Test | p-value:3.59E-04; Z-score:2.91E+00 | ||
|
Methylation in Case |
2.99E-01 (Median) | Methylation in Control | 2.08E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC11A1 in lung adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg14645545) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.47E+00 | Statistic Test | p-value:4.41E-03; Z-score:2.78E+00 | ||
|
Methylation in Case |
3.52E-01 (Median) | Methylation in Control | 2.39E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC11A1 in lung adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg04337188) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:7.99E-03; Z-score:-1.72E+00 | ||
|
Methylation in Case |
7.85E-01 (Median) | Methylation in Control | 8.18E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC11A1 in lung adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg07015784) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:2.05E-02; Z-score:-3.06E+00 | ||
|
Methylation in Case |
9.10E-01 (Median) | Methylation in Control | 9.37E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC11A1 in lung adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg03710889) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.16E+00 | Statistic Test | p-value:3.76E-02; Z-score:1.41E+00 | ||
|
Methylation in Case |
6.52E-01 (Median) | Methylation in Control | 5.63E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC11A1 in lung adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg17010118) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.20E-02; Z-score:-6.00E-01 | ||
|
Methylation in Case |
8.93E-01 (Median) | Methylation in Control | 9.00E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC11A1 in panic disorder | [ 8 ] | |||
|
Location |
TSS1500 (cg13802355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:9.49E-01 | Statistic Test | p-value:2.30E-02; Z-score:3.36E-01 | ||
|
Methylation in Case |
-1.09E+00 (Median) | Methylation in Control | -1.14E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC11A1 in panic disorder | [ 8 ] | |||
|
Location |
Body (cg00634542) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.38E+00 | Statistic Test | p-value:7.71E-03; Z-score:-4.00E-01 | ||
|
Methylation in Case |
5.34E-01 (Median) | Methylation in Control | 7.39E-01 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC11A1 in panic disorder | [ 8 ] | |||
|
Location |
Body (cg04656051) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-8.98E-01 | Statistic Test | p-value:1.19E-02; Z-score:-4.29E-01 | ||
|
Methylation in Case |
-2.20E+00 (Median) | Methylation in Control | -1.98E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC11A1 in panic disorder | [ 8 ] | |||
|
Location |
3'UTR (cg24007161) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:9.82E-01 | Statistic Test | p-value:4.83E-02; Z-score:3.39E-01 | ||
|
Methylation in Case |
-4.97E+00 (Median) | Methylation in Control | -5.07E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
13 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC11A1 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
TSS1500 (cg15099037) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.43E+00 | Statistic Test | p-value:3.34E-16; Z-score:-3.70E+00 | ||
|
Methylation in Case |
4.61E-01 (Median) | Methylation in Control | 6.59E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC11A1 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
TSS200 (cg07719512) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.79E-02; Z-score:-5.16E-01 | ||
|
Methylation in Case |
6.38E-01 (Median) | Methylation in Control | 6.61E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC11A1 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
Body (cg01520853) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:6.93E-05; Z-score:-7.92E-01 | ||
|
Methylation in Case |
9.20E-01 (Median) | Methylation in Control | 9.31E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC11A1 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
Body (cg04656051) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.16E+00 | Statistic Test | p-value:7.63E-05; Z-score:9.77E-01 | ||
|
Methylation in Case |
3.62E-01 (Median) | Methylation in Control | 3.13E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC11A1 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
Body (cg17130789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:2.01E-04; Z-score:-7.86E-01 | ||
|
Methylation in Case |
8.46E-01 (Median) | Methylation in Control | 8.67E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC11A1 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
Body (cg11378979) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.45E-04; Z-score:-6.58E-01 | ||
|
Methylation in Case |
9.02E-01 (Median) | Methylation in Control | 9.13E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC11A1 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
Body (cg14645545) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:3.93E-04; Z-score:7.05E-01 | ||
|
Methylation in Case |
3.27E-01 (Median) | Methylation in Control | 2.87E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC11A1 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
Body (cg03291396) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.16E+00 | Statistic Test | p-value:6.28E-04; Z-score:6.60E-01 | ||
|
Methylation in Case |
2.16E-01 (Median) | Methylation in Control | 1.86E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC11A1 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
Body (cg00634542) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:9.11E-04; Z-score:-8.01E-01 | ||
|
Methylation in Case |
9.27E-01 (Median) | Methylation in Control | 9.41E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC11A1 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
Body (cg03710889) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:2.45E-03; Z-score:9.53E-01 | ||
|
Methylation in Case |
6.77E-01 (Median) | Methylation in Control | 6.11E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC11A1 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
Body (cg23212579) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:3.24E-03; Z-score:-2.29E-01 | ||
|
Methylation in Case |
9.65E-01 (Median) | Methylation in Control | 9.66E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC11A1 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
Body (cg16926316) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.16E-02; Z-score:-3.08E-01 | ||
|
Methylation in Case |
8.48E-01 (Median) | Methylation in Control | 8.53E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC11A1 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
3'UTR (cg24007161) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.17E+00 | Statistic Test | p-value:8.88E-05; Z-score:-7.98E-01 | ||
|
Methylation in Case |
4.14E-02 (Median) | Methylation in Control | 4.82E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC11A1 in prostate cancer | [ 10 ] | |||
|
Location |
TSS200 (cg02595219) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.43E+00 | Statistic Test | p-value:1.01E-03; Z-score:1.22E+01 | ||
|
Methylation in Case |
7.45E-01 (Median) | Methylation in Control | 3.06E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC11A1 in prostate cancer | [ 10 ] | |||
|
Location |
TSS200 (cg09408768) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.22E+00 | Statistic Test | p-value:1.48E-02; Z-score:8.03E+00 | ||
|
Methylation in Case |
3.07E-01 (Median) | Methylation in Control | 1.38E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC11A1 in prostate cancer | [ 10 ] | |||
|
Location |
Body (cg19854293) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.27E+00 | Statistic Test | p-value:8.84E-03; Z-score:3.63E+00 | ||
|
Methylation in Case |
8.33E-01 (Median) | Methylation in Control | 6.53E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC11A1 in clear cell renal cell carcinoma | [ 11 ] | |||
|
Location |
Body (cg00634542) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:6.62E-07; Z-score:-3.05E+00 | ||
|
Methylation in Case |
9.13E-01 (Median) | Methylation in Control | 9.56E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC11A1 in clear cell renal cell carcinoma | [ 11 ] | |||
|
Location |
Body (cg14645545) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.41E+00 | Statistic Test | p-value:6.98E-06; Z-score:1.93E+00 | ||
|
Methylation in Case |
2.10E-01 (Median) | Methylation in Control | 8.72E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC11A1 in clear cell renal cell carcinoma | [ 11 ] | |||
|
Location |
Body (cg04337188) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.55E-05; Z-score:-1.58E+00 | ||
|
Methylation in Case |
9.00E-01 (Median) | Methylation in Control | 9.29E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC11A1 in clear cell renal cell carcinoma | [ 11 ] | |||
|
Location |
Body (cg03291396) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.91E+00 | Statistic Test | p-value:1.82E-05; Z-score:1.01E+00 | ||
|
Methylation in Case |
1.26E-01 (Median) | Methylation in Control | 6.57E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC11A1 in clear cell renal cell carcinoma | [ 11 ] | |||
|
Location |
Body (cg17010118) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.78E-04; Z-score:-1.52E+00 | ||
|
Methylation in Case |
9.74E-01 (Median) | Methylation in Control | 9.80E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC11A1 in clear cell renal cell carcinoma | [ 11 ] | |||
|
Location |
Body (cg17130789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:3.72E-04; Z-score:-1.41E+00 | ||
|
Methylation in Case |
8.78E-01 (Median) | Methylation in Control | 8.99E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC11A1 in clear cell renal cell carcinoma | [ 11 ] | |||
|
Location |
Body (cg07015784) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.00E+00 | Statistic Test | p-value:2.28E-02; Z-score:1.70E-02 | ||
|
Methylation in Case |
9.64E-01 (Median) | Methylation in Control | 9.64E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC11A1 in clear cell renal cell carcinoma | [ 11 ] | |||
|
Location |
Body (cg16926316) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.33E-02; Z-score:-5.92E-01 | ||
|
Methylation in Case |
9.10E-01 (Median) | Methylation in Control | 9.19E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC11A1 in clear cell renal cell carcinoma | [ 11 ] | |||
|
Location |
3'UTR (cg01379701) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:6.11E-03; Z-score:2.44E-01 | ||
|
Methylation in Case |
1.87E-02 (Median) | Methylation in Control | 1.80E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Obesity |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypermethylation of SLC11A1 in obesity than that in healthy individual | ||||
Studied Phenotype |
Obesity [ICD-11:5B81] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:7.27E-20; Fold-change:0.277008523; Z-score:2.907798559 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
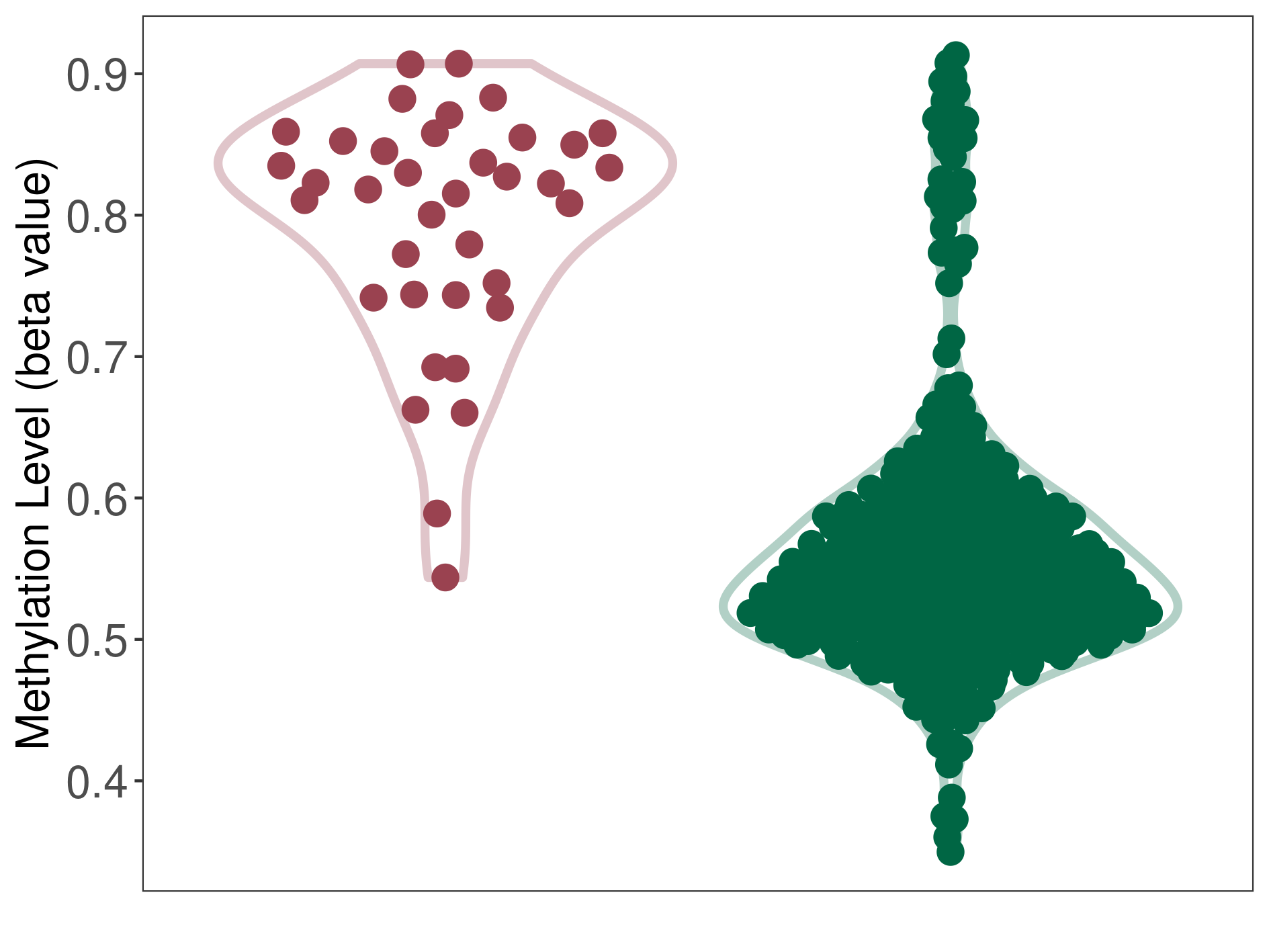
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Anaplastic pilocytic astrocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC11A1 in anaplastic pilocytic astrocytoma than that in healthy individual | ||||
Studied Phenotype |
Anaplastic pilocytic astrocytoma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:8.14E-08; Fold-change:-0.218390547; Z-score:-1.416137848 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
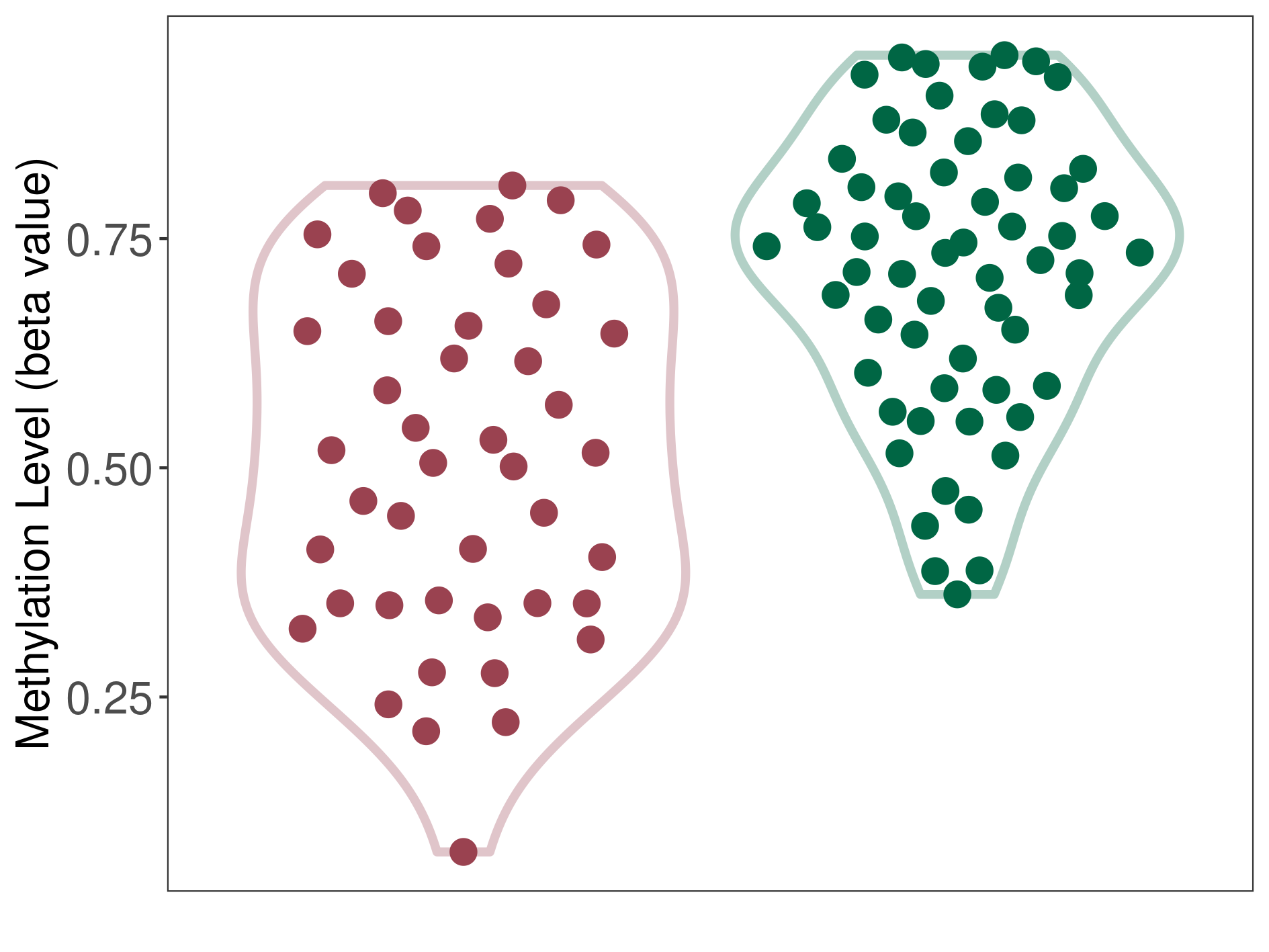
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Anaplastic pleomorphic xanthoastrocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC11A1 in anaplastic pleomorphic xanthoastrocytoma than that in healthy individual | ||||
Studied Phenotype |
Anaplastic pleomorphic xanthoastrocytoma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.001164993; Fold-change:-0.23154836; Z-score:-1.299217618 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
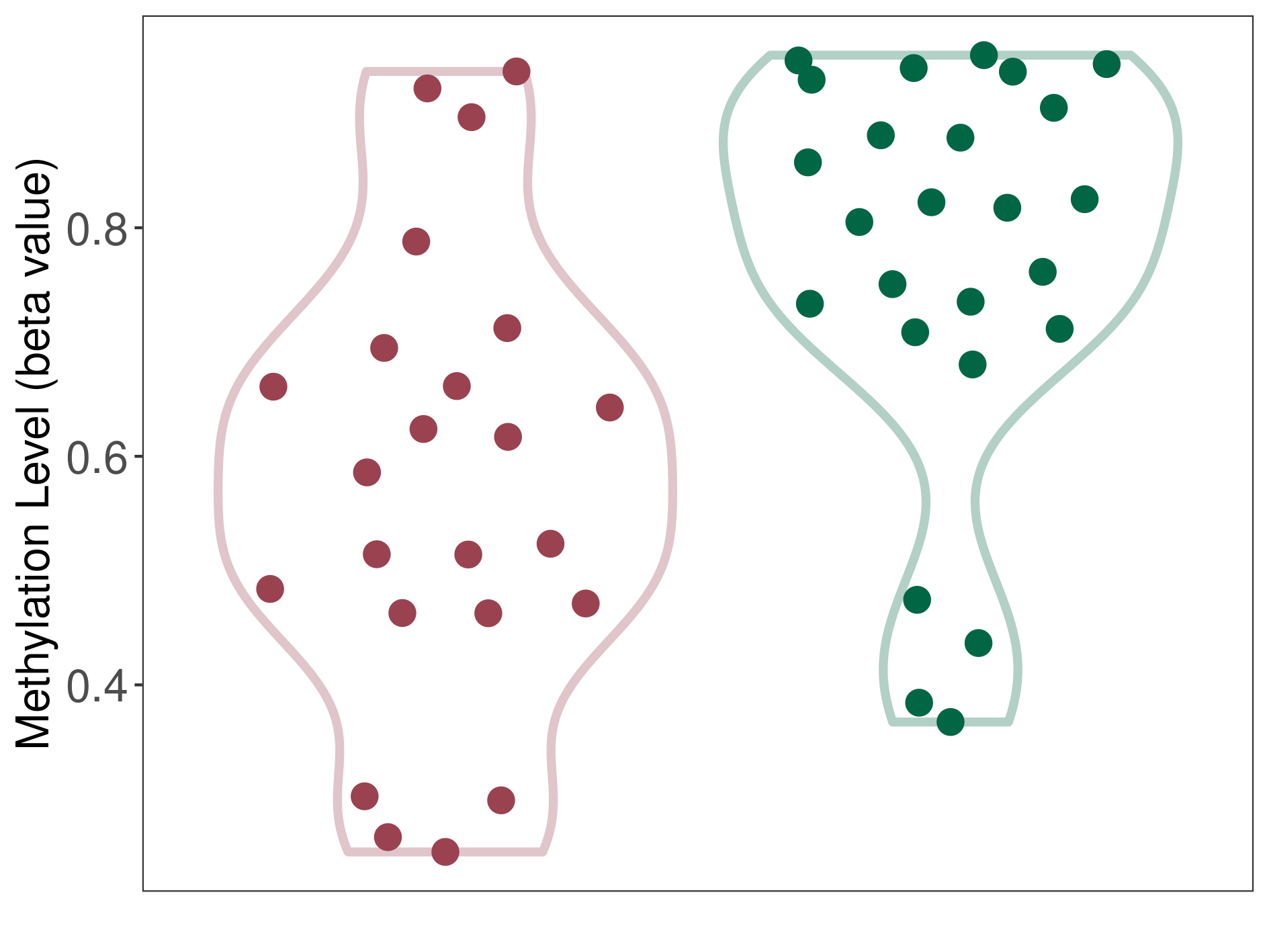
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Cerebral hemispheric glioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC11A1 in cerebral hemispheric glioma than that in healthy individual | ||||
Studied Phenotype |
Cerebral hemispheric glioma [ICD-11:2A00.5] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.00128614; Fold-change:-0.28786962; Z-score:-1.711180215 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
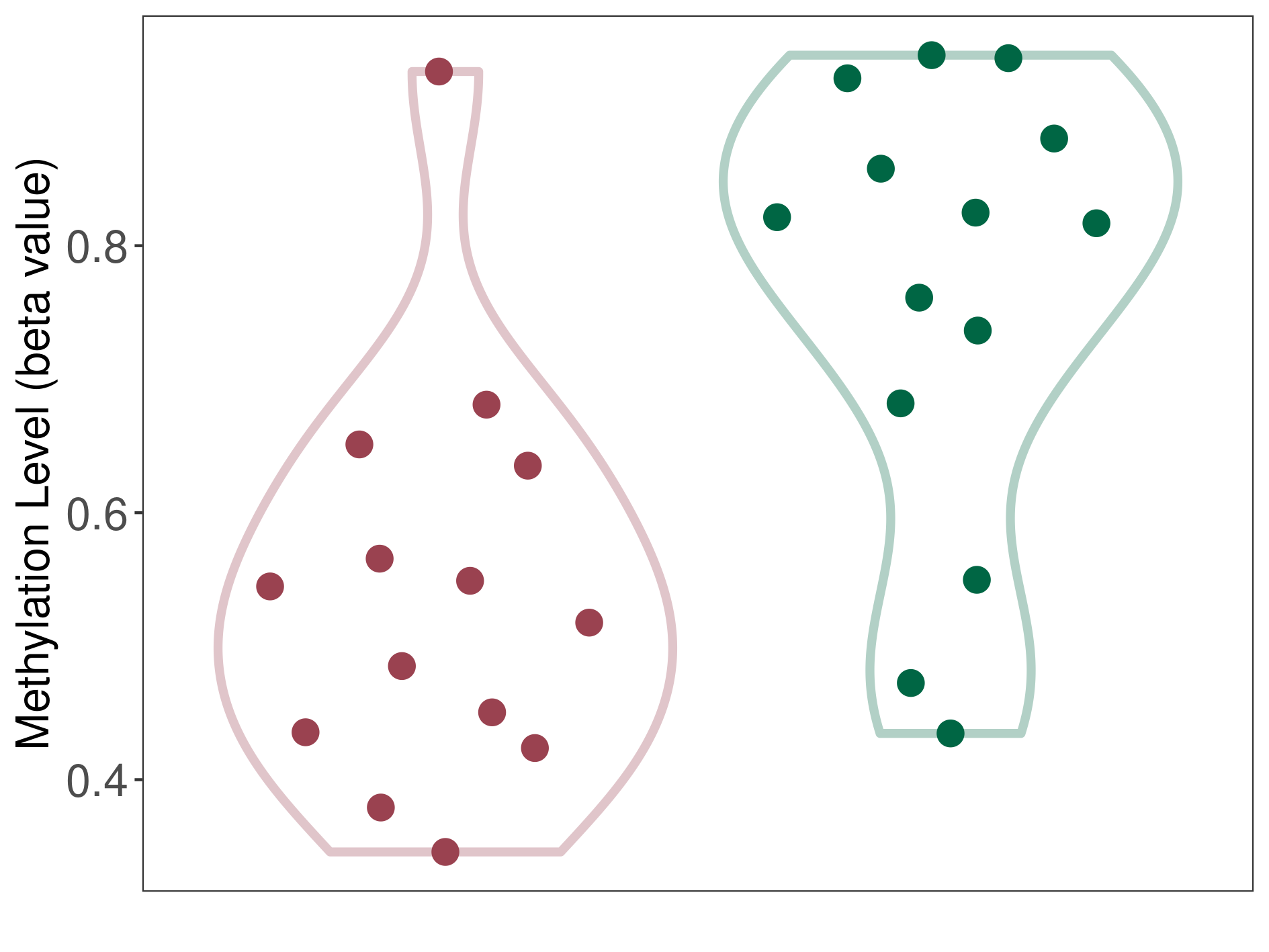
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Chordoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC11A1 in chordoma than that in healthy individual | ||||
Studied Phenotype |
Chordoma [ICD-11:5A61.0] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.000724679; Fold-change:-0.279618904; Z-score:-2.474746527 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
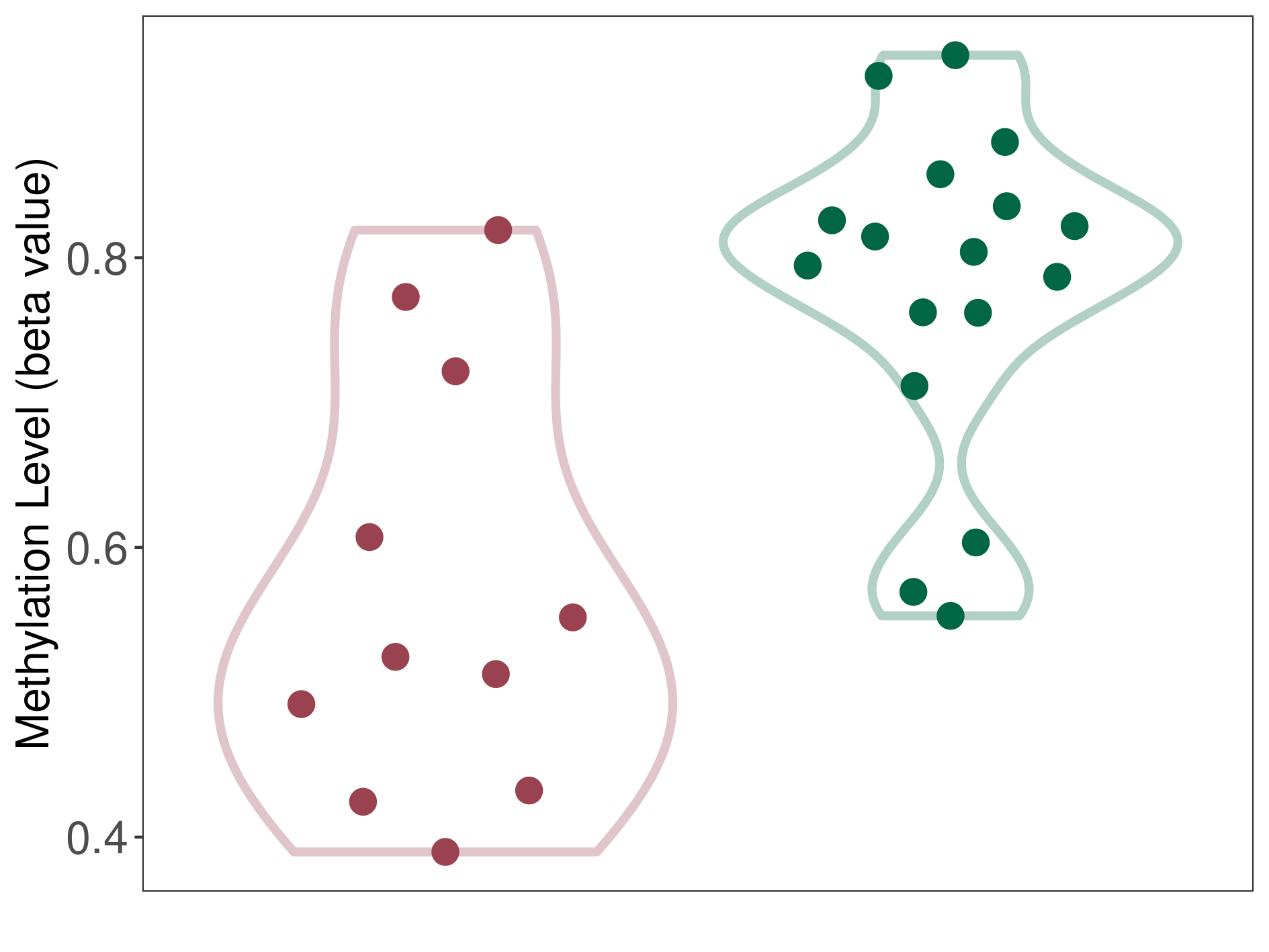
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Craniopharyngioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC11A1 in craniopharyngioma than that in healthy individual | ||||
Studied Phenotype |
Craniopharyngioma [ICD-11:2F9A] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:2.12E-05; Fold-change:-0.22221579; Z-score:-1.325712884 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
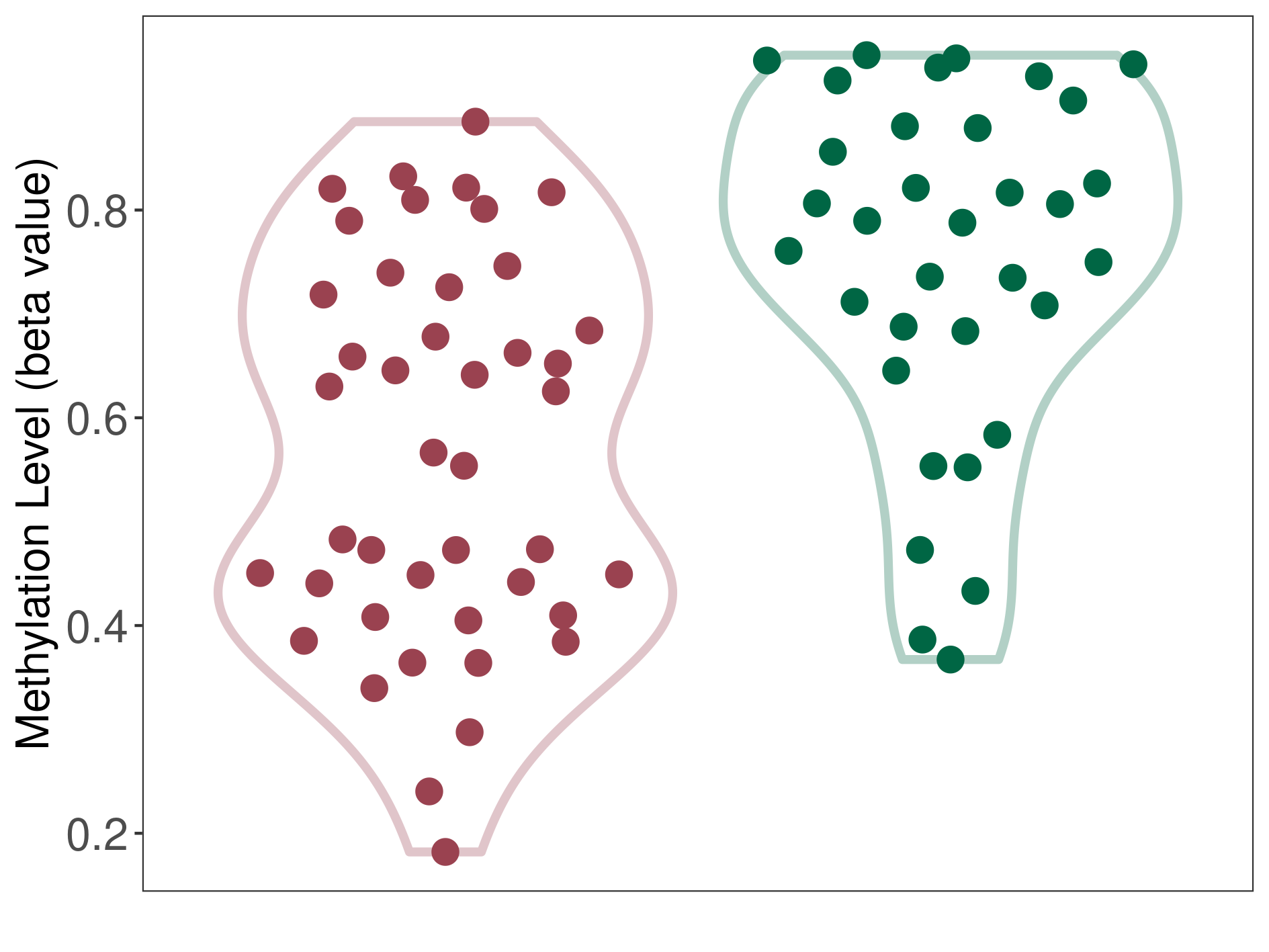
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Diffuse midline glioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC11A1 in diffuse midline glioma than that in healthy individual | ||||
Studied Phenotype |
Diffuse midline glioma [ICD-11:2A00.0Z] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:7.27E-19; Fold-change:-0.252639361; Z-score:-1.397814588 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
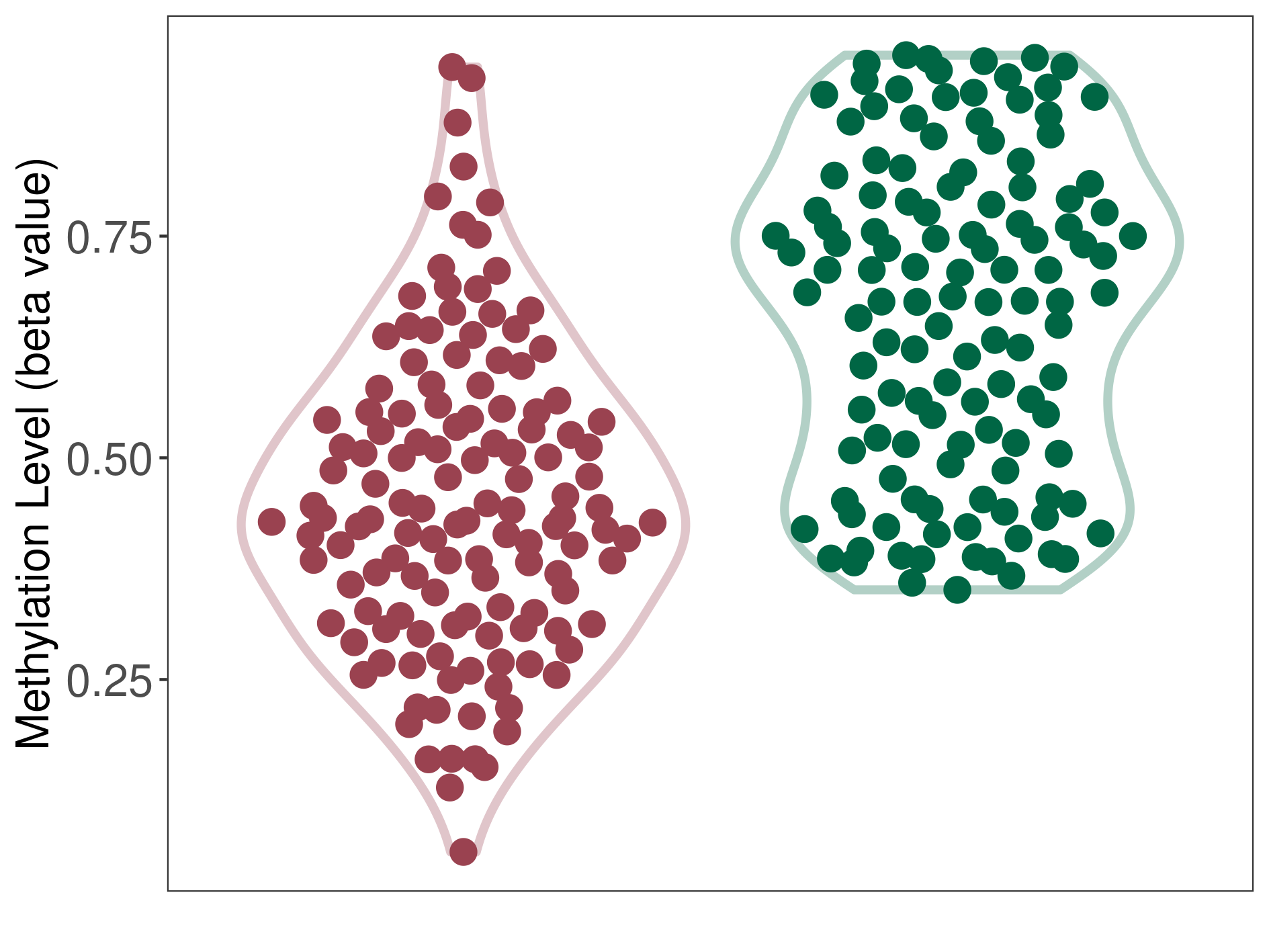
|
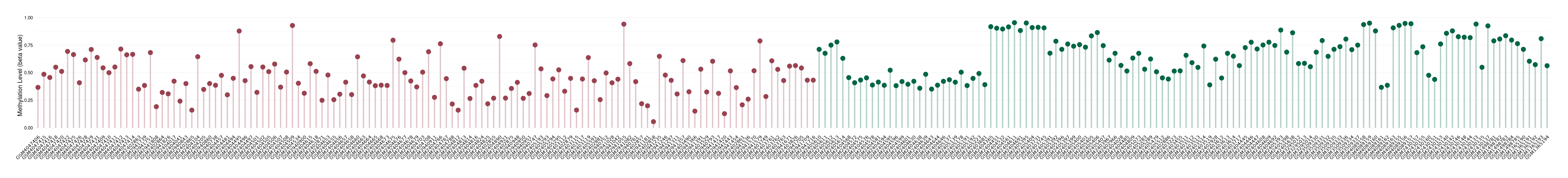 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
RELA YAP fusion ependymoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC11A1 in rela yap fusion ependymoma than that in healthy individual | ||||
Studied Phenotype |
RELA YAP fusion ependymoma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:9.69E-11; Fold-change:-0.21659148; Z-score:-1.361411235 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
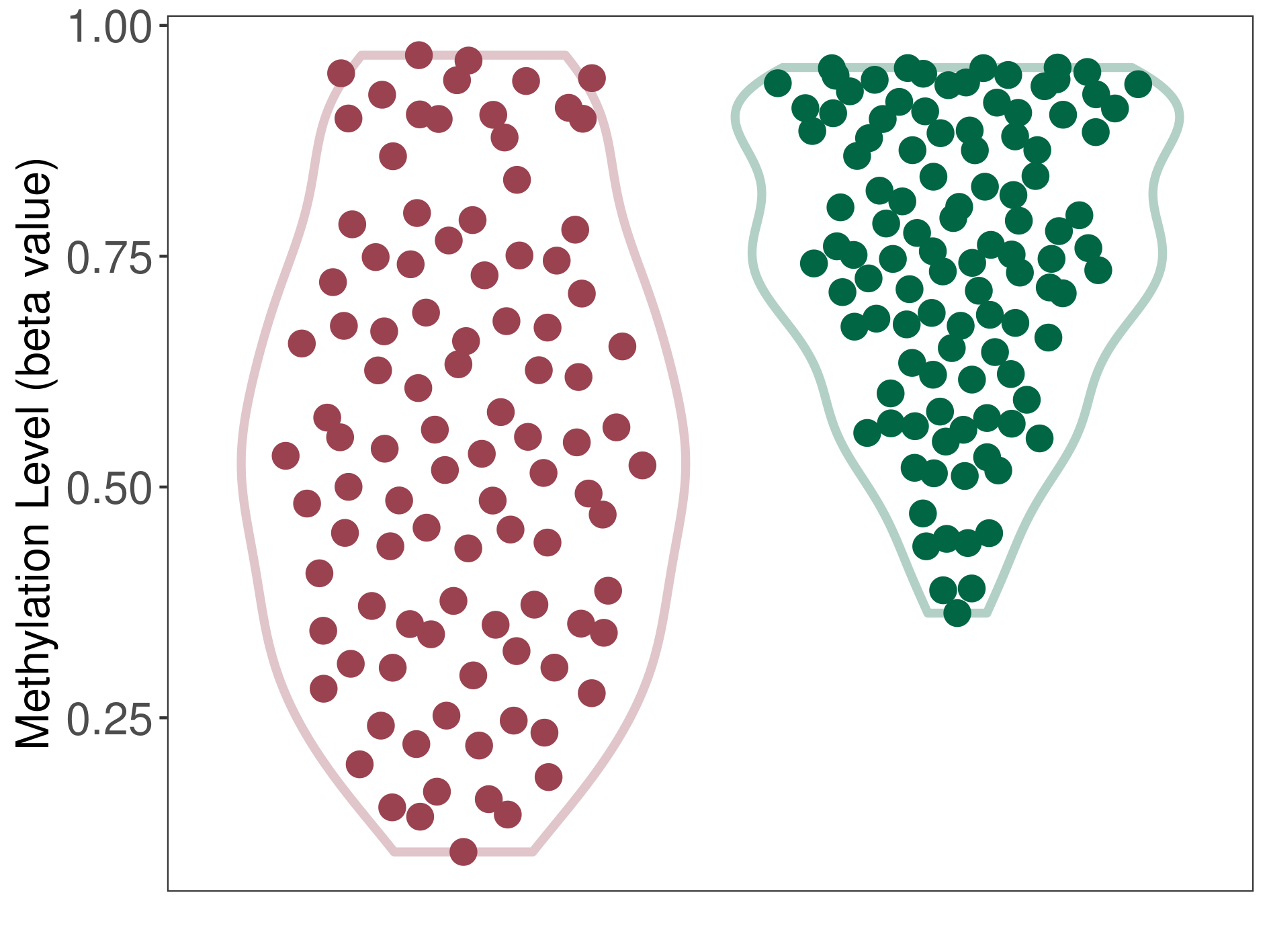
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Brain neuroblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC11A1 in brain neuroblastoma than that in healthy individual | ||||
Studied Phenotype |
Brain neuroblastoma [ICD-11:2A00.11] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.33E-13; Fold-change:-0.374098079; Z-score:-2.355102323 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
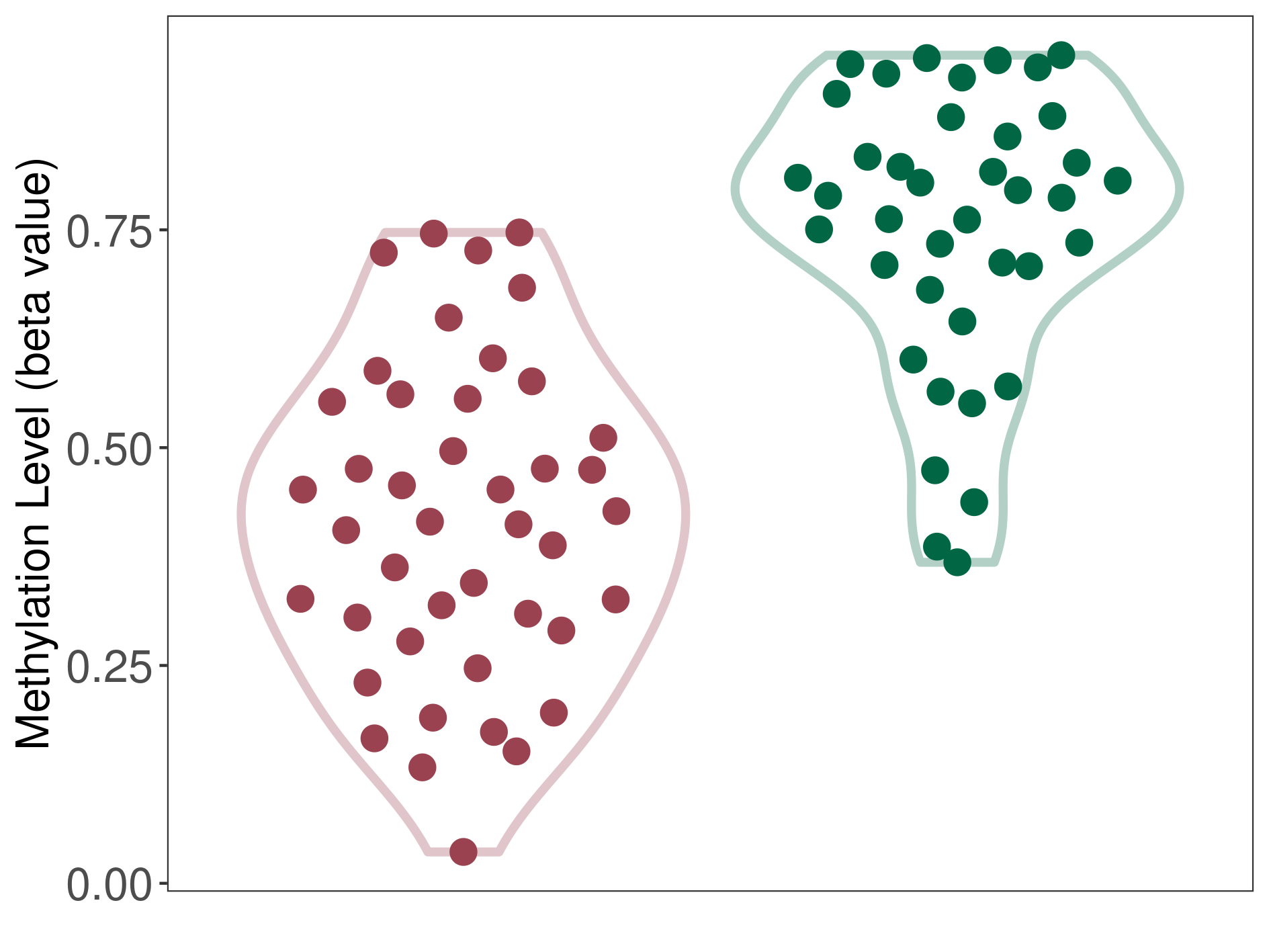
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Brain neuroepithelial tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC11A1 in brain neuroepithelial tumour than that in healthy individual | ||||
Studied Phenotype |
Brain neuroepithelial tumour [ICD-11:2A00.2Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.66E-13; Fold-change:-0.442569994; Z-score:-2.875966396 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
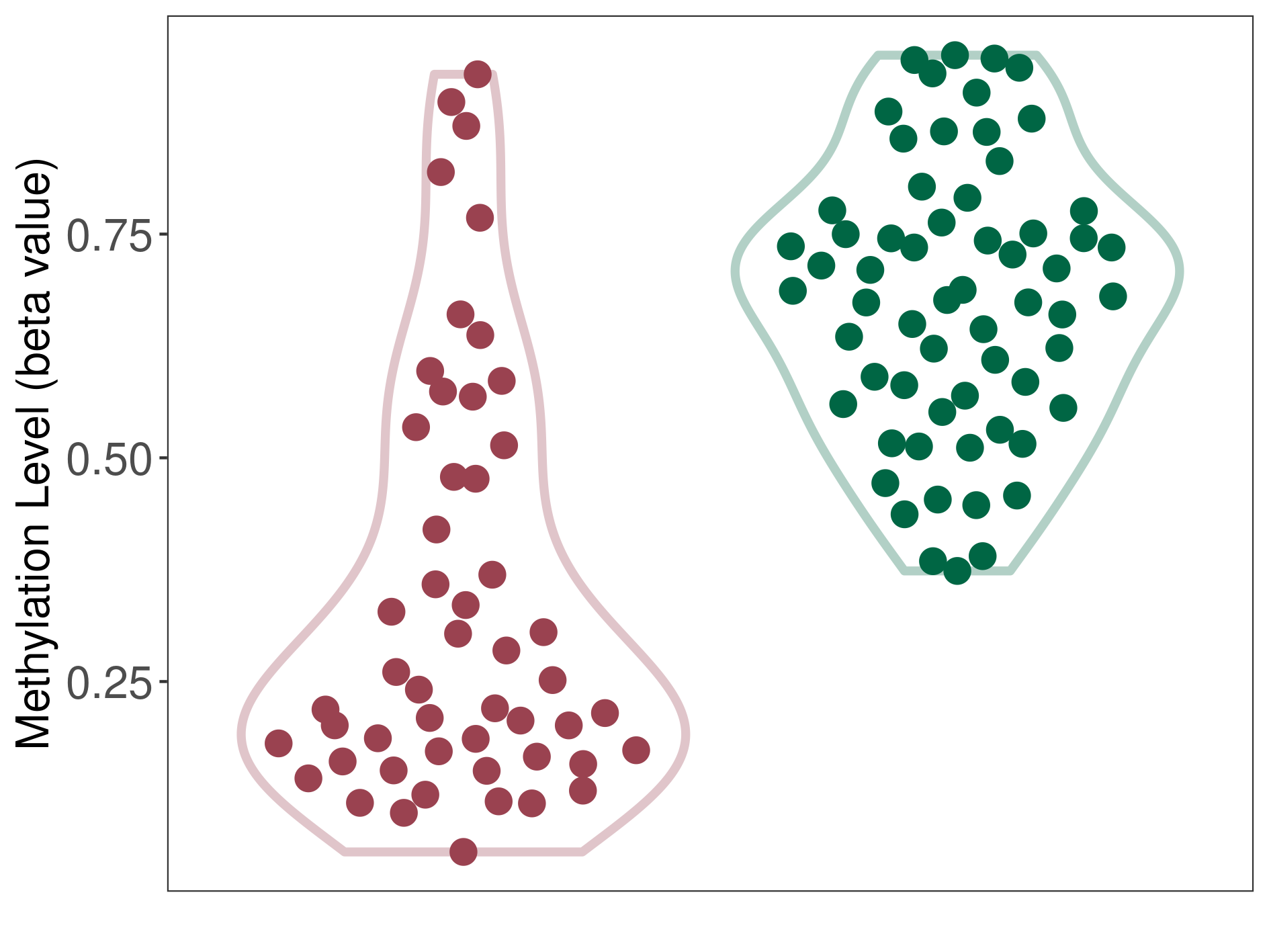
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Cerebellar liponeurocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC11A1 in cerebellar liponeurocytoma than that in healthy individual | ||||
Studied Phenotype |
Cerebellar liponeurocytoma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:3.33E-10; Fold-change:-0.59127381; Z-score:-3.485579053 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
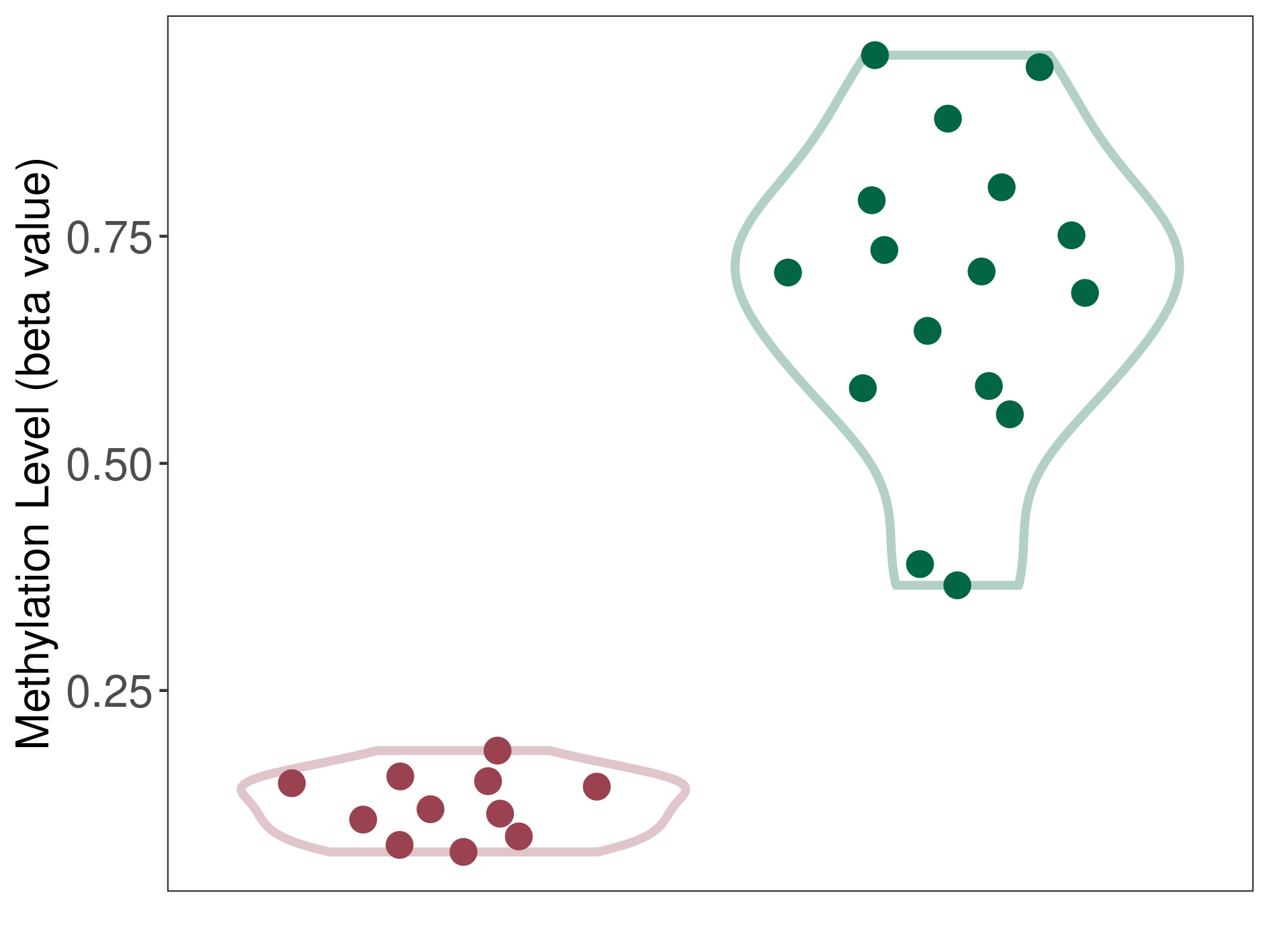
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Glioblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC11A1 in glioblastoma than that in healthy individual | ||||
Studied Phenotype |
Glioblastoma [ICD-11:2A00.00] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:6.50E-13; Fold-change:-0.3668635; Z-score:-2.131240355 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
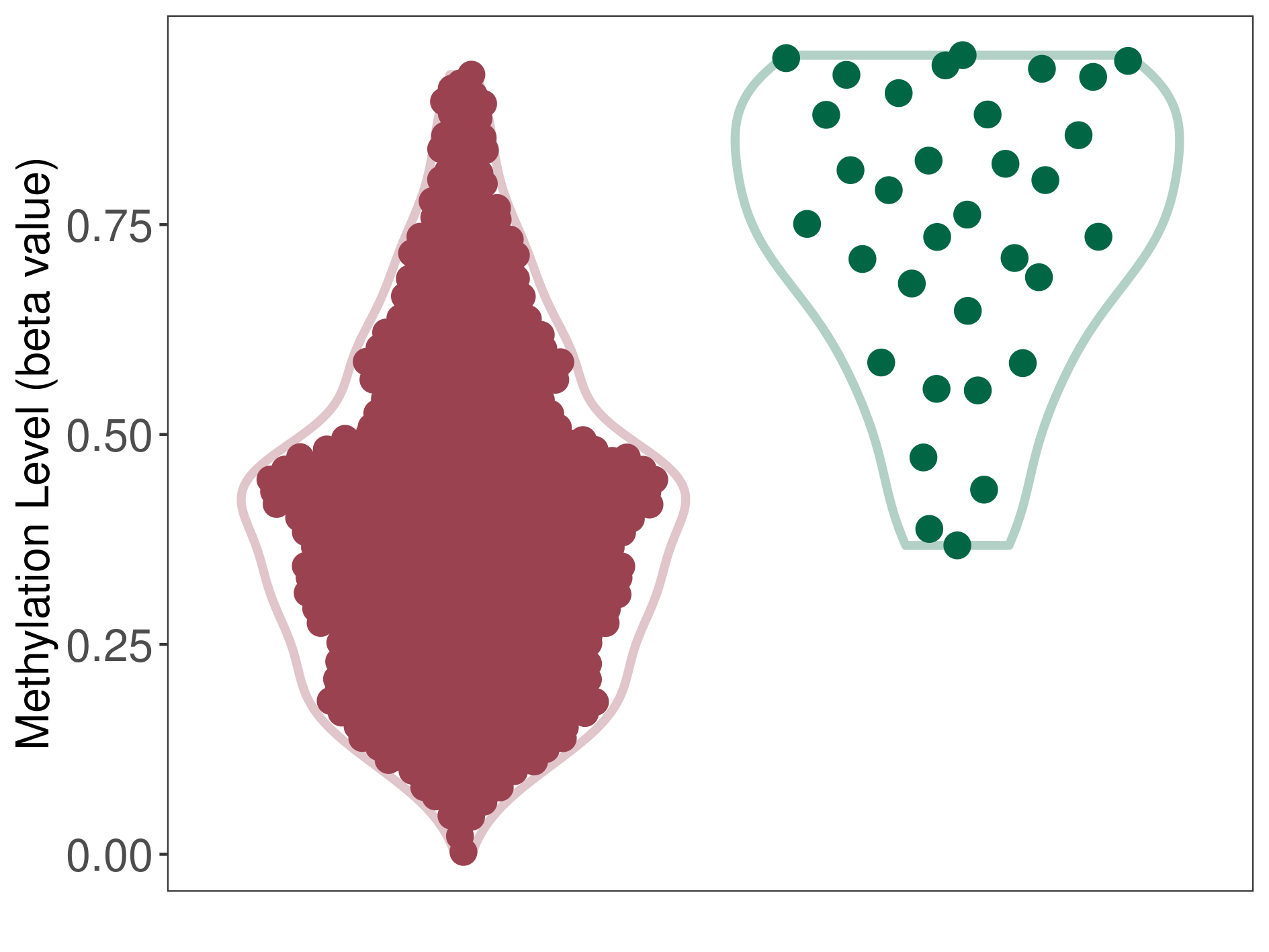
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Lymphoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC11A1 in lymphoma than that in healthy individual | ||||
Studied Phenotype |
Lymphoma [ICD-11:2B30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:7.26E-33; Fold-change:-0.597484731; Z-score:-3.960907715 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
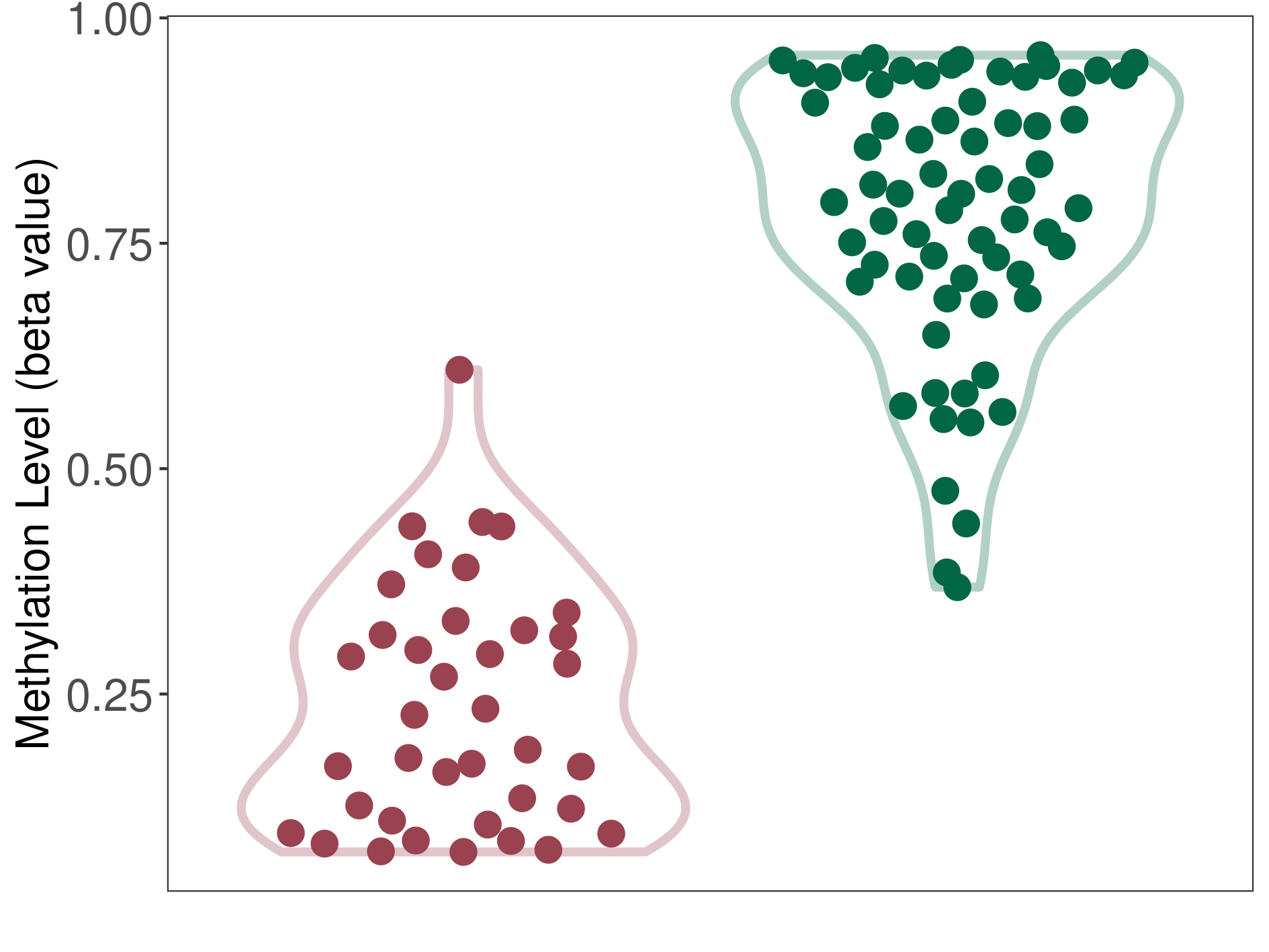
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Multilayered rosettes embryonal tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC11A1 in multilayered rosettes embryonal tumour than that in healthy individual | ||||
Studied Phenotype |
Multilayered rosettes embryonal tumour [ICD-11:2A00.1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.32E-22; Fold-change:-0.512063328; Z-score:-3.287101202 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
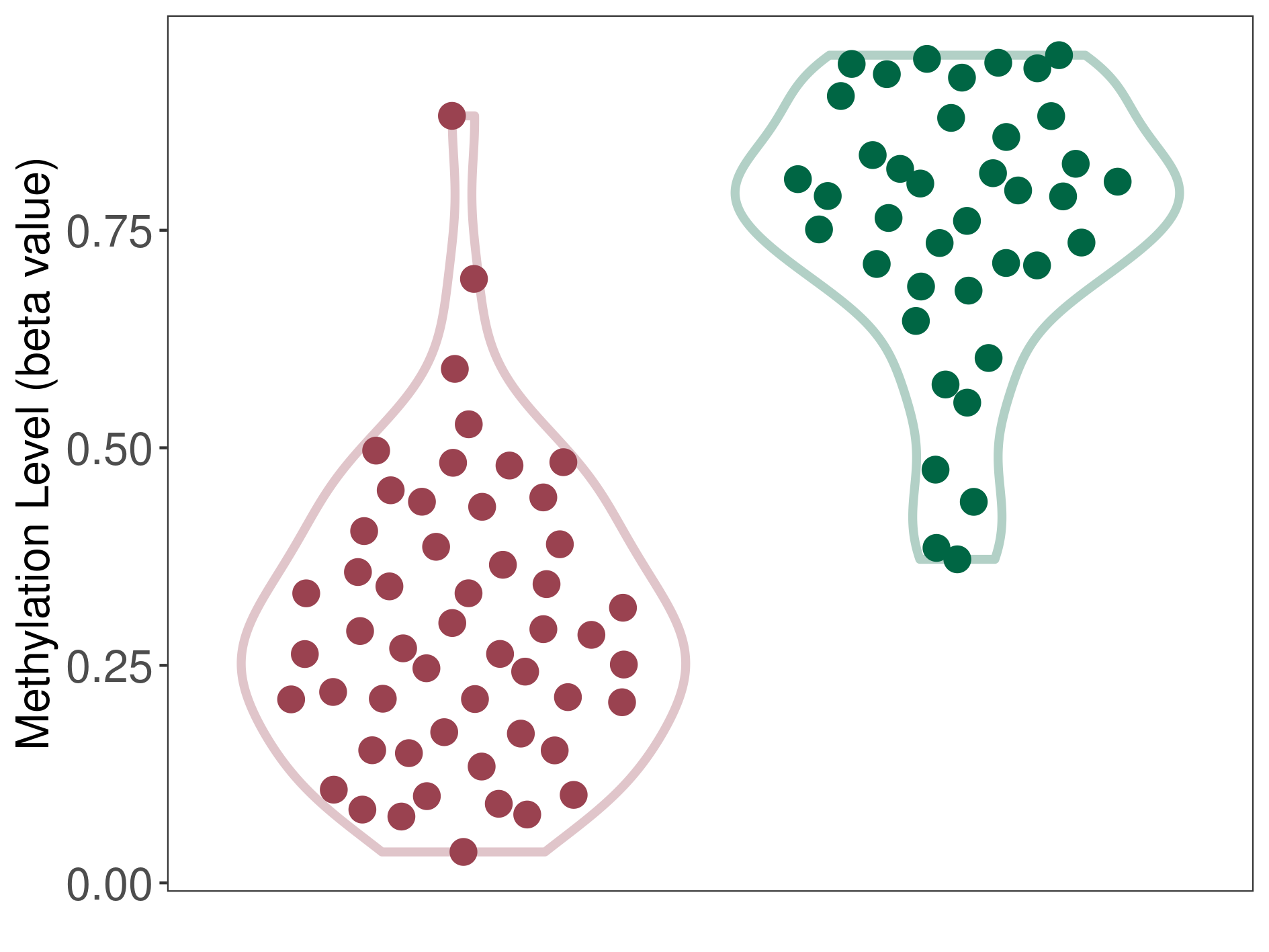
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Posterior fossa ependymoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC11A1 in posterior fossa ependymoma than that in healthy individual | ||||
Studied Phenotype |
Posterior fossa ependymoma [ICD-11:2D50.2] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.00E-86; Fold-change:-0.57869342; Z-score:-3.242845843 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
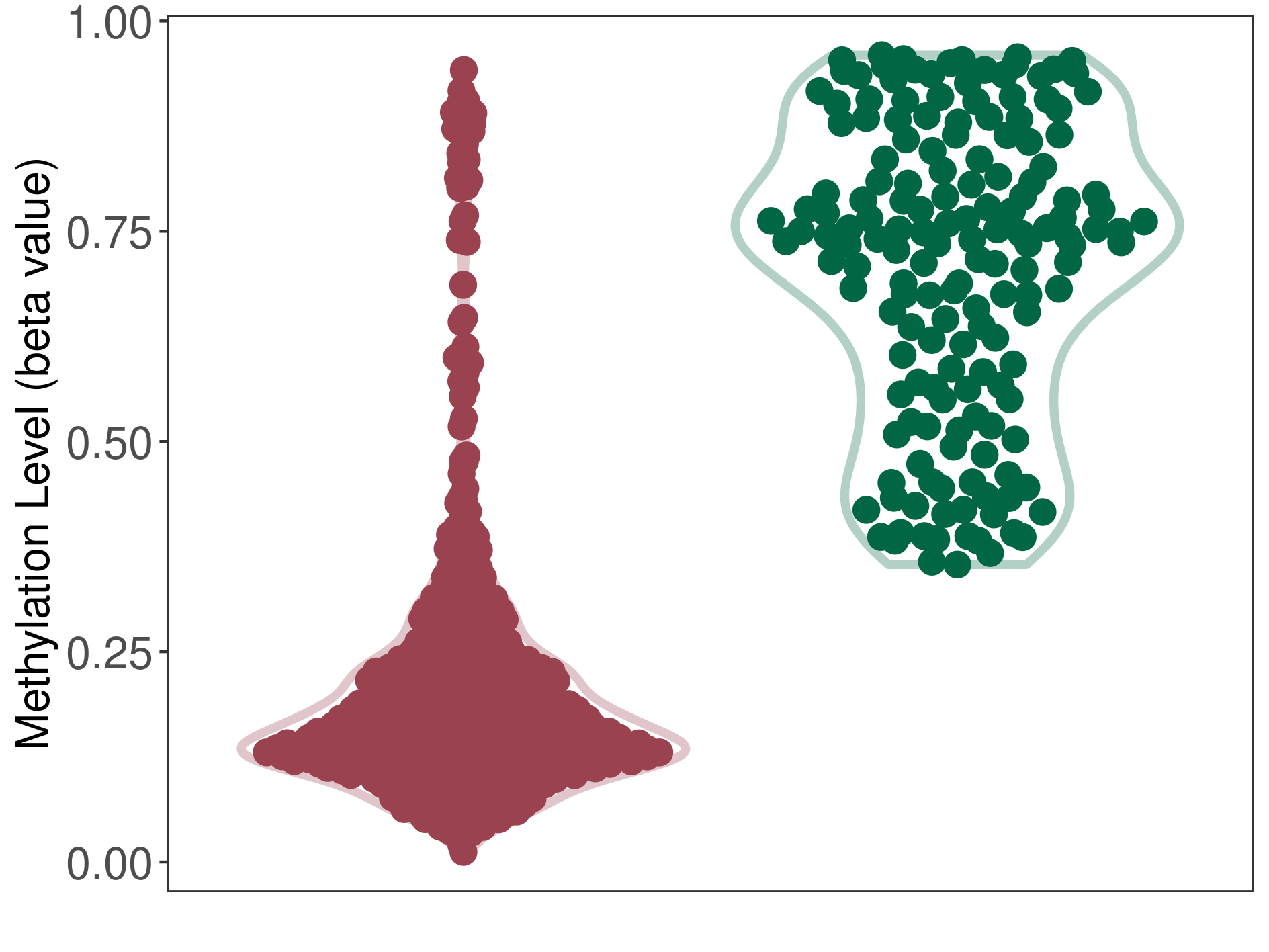
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
microRNA |
|||||
|
Unclear Phenotype |
18 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
miR-1199 directly targets SLC11A1 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1199 | miRNA Mature ID | miR-1199-3p | ||
|
miRNA Sequence |
UGCGGCCGGUGCUCAACCUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon2 |
miR-1233 directly targets SLC11A1 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1233 | miRNA Mature ID | miR-1233-5p | ||
|
miRNA Sequence |
AGUGGGAGGCCAGGGCACGGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon3 |
miR-182 directly targets SLC11A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-182 | miRNA Mature ID | miR-182-5p | ||
|
miRNA Sequence |
UUUGGCAAUGGUAGAACUCACACU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon4 |
miR-1908 directly targets SLC11A1 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1908 | miRNA Mature ID | miR-1908-3p | ||
|
miRNA Sequence |
CCGGCCGCCGGCUCCGCCCCG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon5 |
miR-196a directly targets SLC11A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-196a | miRNA Mature ID | miR-196a-3p | ||
|
miRNA Sequence |
CGGCAACAAGAAACUGCCUGAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon6 |
miR-339 directly targets SLC11A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-339 | miRNA Mature ID | miR-339-5p | ||
|
miRNA Sequence |
UCCCUGUCCUCCAGGAGCUCACG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon7 |
miR-4446 directly targets SLC11A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4446 | miRNA Mature ID | miR-4446-5p | ||
|
miRNA Sequence |
AUUUCCCUGCCAUUCCCUUGGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon8 |
miR-4510 directly targets SLC11A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4510 | miRNA Mature ID | miR-4510 | ||
|
miRNA Sequence |
UGAGGGAGUAGGAUGUAUGGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon9 |
miR-4755 directly targets SLC11A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4755 | miRNA Mature ID | miR-4755-5p | ||
|
miRNA Sequence |
UUUCCCUUCAGAGCCUGGCUUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon10 |
miR-5006 directly targets SLC11A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5006 | miRNA Mature ID | miR-5006-3p | ||
|
miRNA Sequence |
UUUCCCUUUCCAUCCUGGCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon11 |
miR-6127 directly targets SLC11A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6127 | miRNA Mature ID | miR-6127 | ||
|
miRNA Sequence |
UGAGGGAGUGGGUGGGAGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon12 |
miR-6129 directly targets SLC11A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6129 | miRNA Mature ID | miR-6129 | ||
|
miRNA Sequence |
UGAGGGAGUUGGGUGUAUA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon13 |
miR-6130 directly targets SLC11A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6130 | miRNA Mature ID | miR-6130 | ||
|
miRNA Sequence |
UGAGGGAGUGGAUUGUAUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon14 |
miR-6133 directly targets SLC11A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6133 | miRNA Mature ID | miR-6133 | ||
|
miRNA Sequence |
UGAGGGAGGAGGUUGGGUA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon15 |
miR-6760 directly targets SLC11A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6760 | miRNA Mature ID | miR-6760-5p | ||
|
miRNA Sequence |
CAGGGAGAAGGUGGAAGUGCAGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon16 |
miR-6778 directly targets SLC11A1 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6778 | miRNA Mature ID | miR-6778-5p | ||
|
miRNA Sequence |
AGUGGGAGGACAGGAGGCAGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon17 |
miR-6853 directly targets SLC11A1 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6853 | miRNA Mature ID | miR-6853-5p | ||
|
miRNA Sequence |
AGCGUGGGAUGUCCAUGAAGUCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon18 |
miR-6873 directly targets SLC11A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6873 | miRNA Mature ID | miR-6873-5p | ||
|
miRNA Sequence |
CAGAGGGAAUACAGAGGGCAAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
