Detail Information of DT Wildtype Structure
| General Information of Drug Transporter (DT) | ||||||
|---|---|---|---|---|---|---|
| DT ID | DTD0081 Transporter Info | |||||
| Gene Name | SLC11A2 | |||||
| Transporter Name | Natural resistance-associated macrophage protein 2 | |||||
| Gene ID | ||||||
| UniProt ID | ||||||
| Modelled Structures of This DT | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Detail Information of Structural Model | ||||||
| Method | Homology modeling | |||||
| Template PDB | 5M94_A | Identity | 36.927% | |||
| Performance | Minimized Score | -1063.606 kcal/mol | ||||
| Ramachandra Favored | Medium |
|||||
| QMEANBrane Quality | High |
|||||
| Ramachandran Plot |
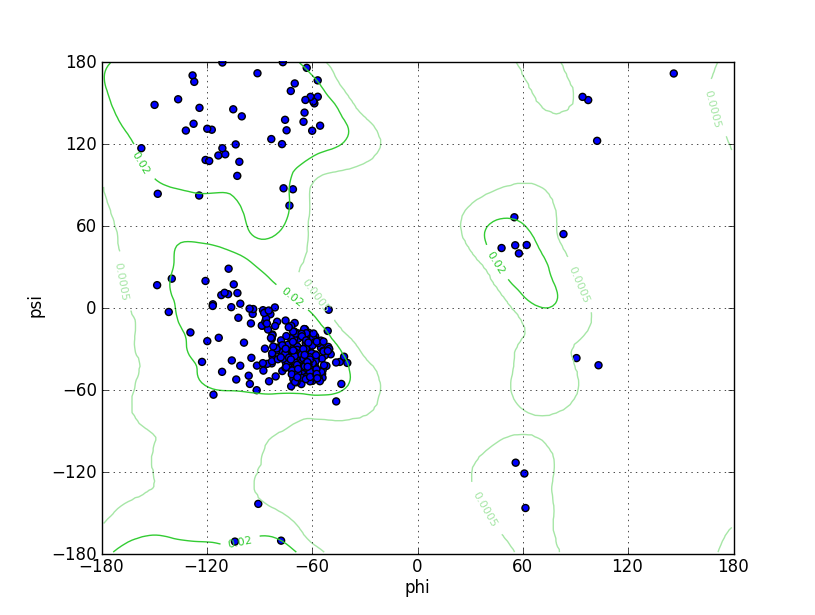
|
|||||
| Click to Save Ramachandran Plot in PNG Format | ||||||
| Ramz Z Score | -1.83 ±0.37 | |||||
| Residues in Favored Region | 406 | Ramachandran favored | 95.08% | |||
| Number of Outliers | 4 | Ramachandran outliers | 0.94% | |||
| Local Quality |
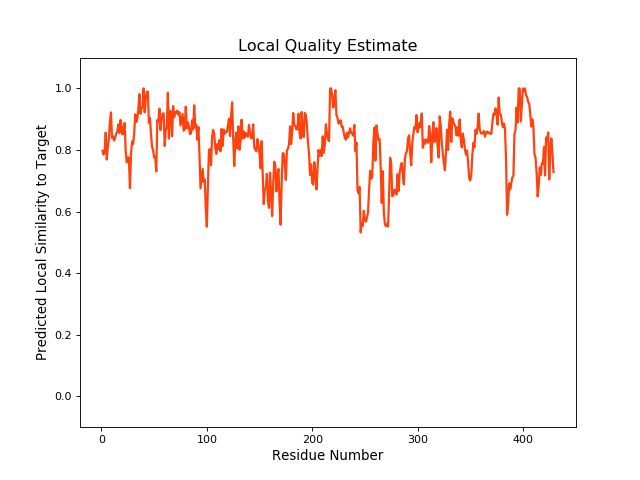
|
|||||
| Click to Save Local Quality Plot in PNG Format | ||||||
| QMEANBrane Score | 0.82 | |||||
| AlphaFold Protein Structure Database | ||||||
| SWISS-MODEL Database | ||||||
| Experimental Structures of This DT | ||||||
| PDB ID | Scanning Method | Resolution | Expression System | Details | Ref | |
| 5F0L | X-ray | 3.2 Å | Escherichia coli BL21(DE3) | [1] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | D | |||||
| Sequence Length | 545-568 | Mutation | No | |||
| 5F0M | X-ray | 3.1 Å | Escherichia coli BL21(DE3); scherichia coli | [1] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | D | |||||
| Sequence Length | 549-560 | Mutation | No | |||
| 5F0P | X-ray | 2.78 Å | Escherichia coli BL21(DE3); scherichia coli | [1] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | D | |||||
| Sequence Length | 549-560 | Mutation | Yes | |||
| 7BLO | EM | 9.5 Å | Escherichia coli | [2] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | H/N | |||||
| Sequence Length | 10 | Mutation | No | |||
| 7BLQ | EM | 9.2 Å | Escherichia coli | [2] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | U/V | |||||
| Sequence Length | 10 | Mutation | No | |||
| 9F6N | EM | 3.6 Å | Homo sapiens | [3] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | A | |||||
| Sequence Length | 655 | Mutation | No | |||
| 9F6O | EM | 3.9 Å | Escherichia coli,Homo sapiens | [3] | ||
| Structure | ||||||
| Click to Save PDB File in TXT Format | ||||||
| Corresponding chain | A | |||||
| Sequence Length | 655 | Mutation | No | |||
| References | ||||||
| 1 | Structural Mechanism for Cargo Recognition by the Retromer Complex. Cell. 2016 Dec 1;167(6):1623-1635.e14. | |||||
| 2 | Architecture and mechanism of metazoan retromer:SNX3 tubular coat assembly. Sci Adv. 2021;7(13). | |||||
| 3 | Structural basis for metal ion transport by the human SLC11 proteins DMT1 and NRAMP1. Nat Commun. 2025;16(1):761. | |||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
