Modelled Stucture
Method:Homology modeling
Teplate PDB:5YKE_B
Identity:67.317%
Minimized Score:-2706.765
Detail: Structure Info
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0062 Transporter Info | ||||
| Gene Name | ABCC9 | ||||
| Protein Name | Sulfonylurea receptor 2 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| 3D Structure |
Modelled Stucture Method:Homology modeling Teplate PDB:5YKE_B Identity:67.317% Minimized Score:-2706.765 Detail: Structure Info |
||||
| Inter-species Structural Differences (ISD) | |||||
| Mus musculus (Mouse) | |||||
| Gene Name | Abcc9 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5YKE_B Sequence Length:1546 Identity:67.19% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -2957.65 kcal/mol | |||
| Ramachandra Favored | Medium |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
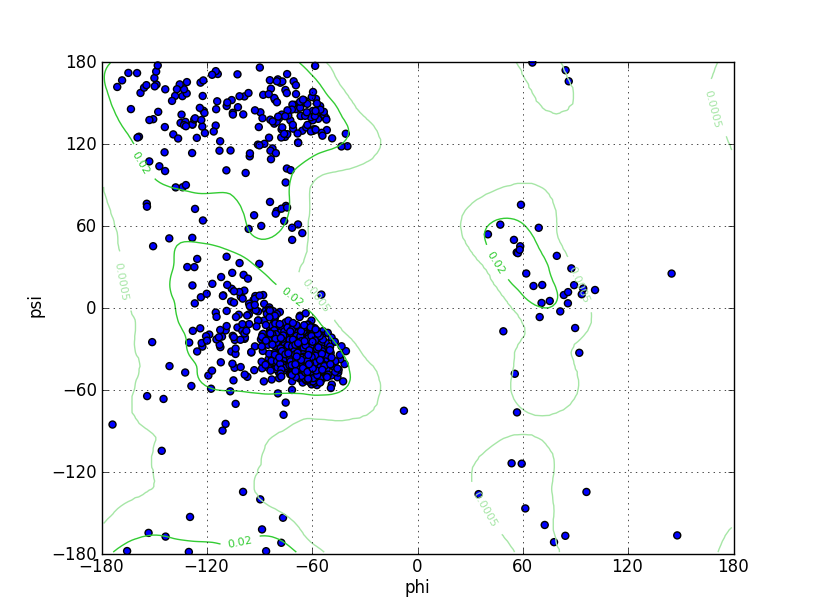
Ramz Z Score:-1.51 ±0.19 Residues in Favored Region:1488 Ramachandran favored:96.37% Number of Outliers:10 Ramachandran outliers:0.65% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
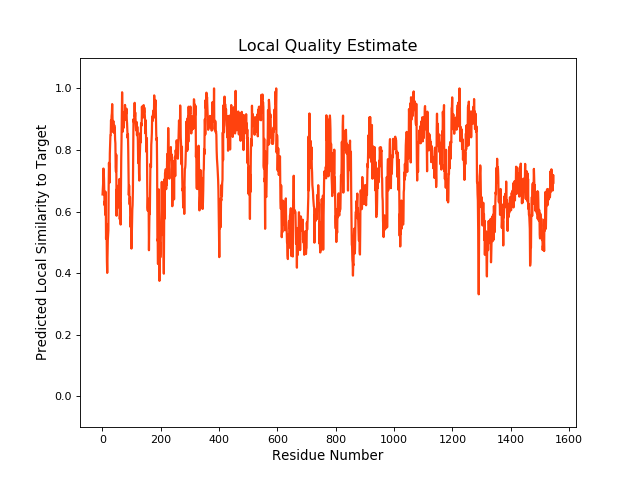
QMEANBrane Score:0.73 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Rattus norvegicus (Rat) | |||||
| Gene Name | Abcc9 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5YKE_B Sequence Length:1545 Identity:67.316% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -2849.781 kcal/mol | |||
| Ramachandra Favored | Medium |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
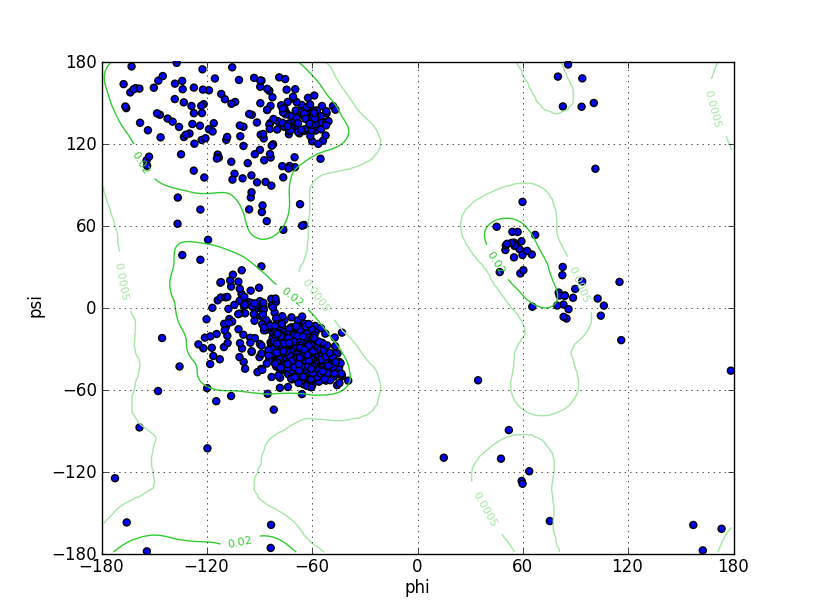
Ramz Z Score:-1.22 ±0.19 Residues in Favored Region:1504 Ramachandran favored:97.47% Number of Outliers:13 Ramachandran outliers:0.84% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
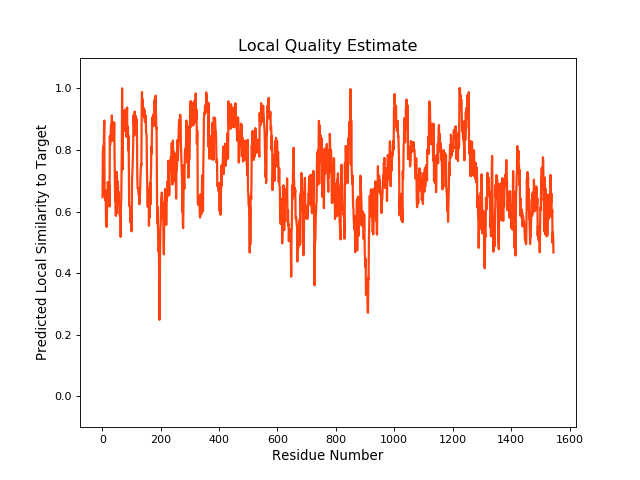
QMEANBrane Score:0.73 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| Oryctolagus cuniculus (Rabbit) | |||||
| Gene Name | ABCC9 | ||||
| UniProt ID | |||||
| UniProt Entry | |||||
| 3D Structure |
Method:Homology modeling Teplate PDB:5YKE_B Sequence Length:1549 Identity:67.254% |
||||
| Click to Save PDB File in TXT Format | |||||
| Performance | Minimized Score | -2849.41 kcal/mol | |||
| Ramachandra Favored | Medium |
||||
| QMEANBrane Quality | Medium |
||||
| Ramachandran Plot |
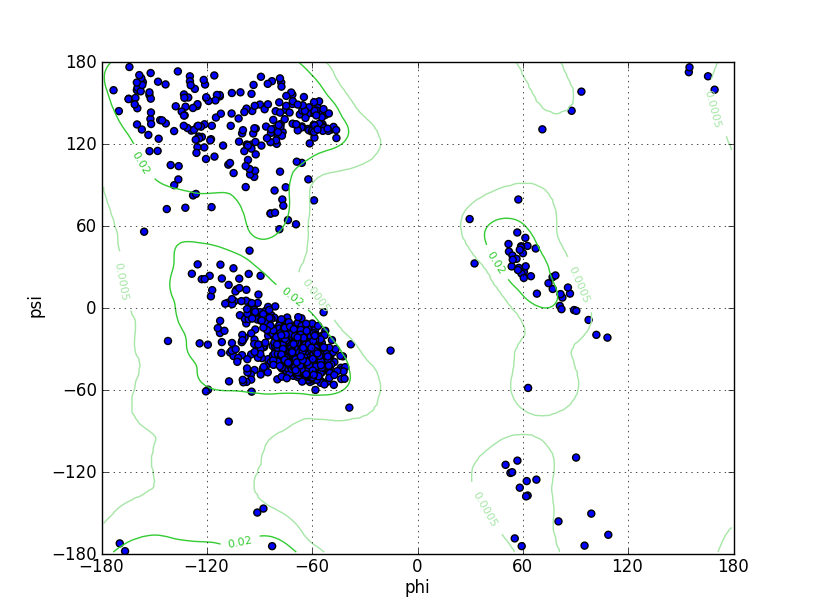
Ramz Z Score:-0.66 ±0.2 Residues in Favored Region:1502 Ramachandran favored:97.09% Number of Outliers:11 Ramachandran outliers:0.71% |
||||
| Click to Save Ramachandran Plot in PNG Format | |||||
| Local Quality |
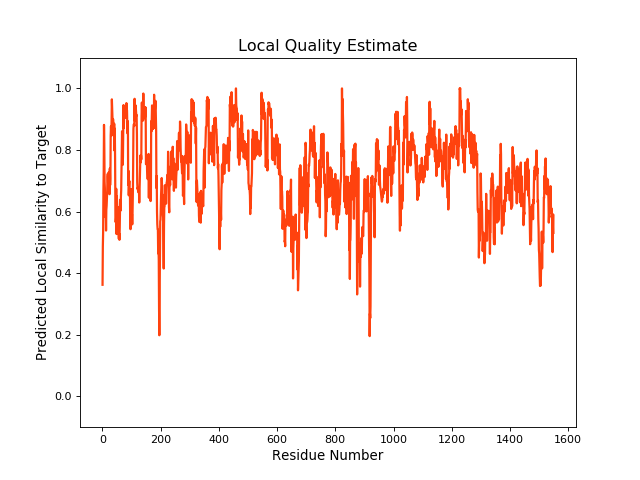
QMEANBrane Score:0.73 |
||||
| Click to Save Local Quality Plot in PNG Format | |||||
| References | |||||
| 1 | Vascular K(ATP) channel structural dynamics reveal regulatory mechanism by Mg-nucleotides. Proc Natl Acad Sci U S A. 2021;118(44). | ||||
| 2 | Structural identification of vasodilator binding sites on the SUR2 subunit. Nat Commun. 2022;13(1):2675. | ||||
| 3 | The inhibition mechanism of the SUR2A-containing K(ATP) channel by a regulatory helix. Nat Commun. 2023;14(1):3608. | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.