Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0477 Transporter Info | ||||
| Gene Name | SLC8A1 | ||||
| Transporter Name | Sodium/calcium exchanger 1 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Bladder cancer |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC8A1 in bladder cancer | [ 1 ] | |||
|
Location |
5'UTR (cg03256780) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.55E+00 | Statistic Test | p-value:4.33E-07; Z-score:-5.36E+00 | ||
|
Methylation in Case |
1.63E-01 (Median) | Methylation in Control | 4.15E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC8A1 in bladder cancer | [ 1 ] | |||
|
Location |
5'UTR (cg08240652) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:6.63E-05; Z-score:-1.07E+01 | ||
|
Methylation in Case |
7.87E-01 (Median) | Methylation in Control | 8.95E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC8A1 in bladder cancer | [ 1 ] | |||
|
Location |
5'UTR (cg15549810) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:5.48E-04; Z-score:-3.13E+00 | ||
|
Methylation in Case |
7.11E-01 (Median) | Methylation in Control | 7.83E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC8A1 in bladder cancer | [ 1 ] | |||
|
Location |
5'UTR (cg19918022) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:2.40E-03; Z-score:-3.80E+00 | ||
|
Methylation in Case |
7.89E-01 (Median) | Methylation in Control | 8.58E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC8A1 in bladder cancer | [ 1 ] | |||
|
Location |
5'UTR (cg00610991) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.77E+00 | Statistic Test | p-value:3.99E-03; Z-score:-3.92E+00 | ||
|
Methylation in Case |
1.29E-01 (Median) | Methylation in Control | 2.29E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC8A1 in bladder cancer | [ 1 ] | |||
|
Location |
5'UTR (cg10633958) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.23E+00 | Statistic Test | p-value:7.71E-03; Z-score:1.68E+00 | ||
|
Methylation in Case |
9.48E-02 (Median) | Methylation in Control | 7.69E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC8A1 in bladder cancer | [ 1 ] | |||
|
Location |
TSS1500 (cg02870829) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.61E+00 | Statistic Test | p-value:3.83E-08; Z-score:-6.80E+00 | ||
|
Methylation in Case |
4.26E-01 (Median) | Methylation in Control | 6.87E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC8A1 in bladder cancer | [ 1 ] | |||
|
Location |
TSS1500 (cg02577240) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:8.50E-04; Z-score:-3.79E+00 | ||
|
Methylation in Case |
5.95E-01 (Median) | Methylation in Control | 8.04E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC8A1 in bladder cancer | [ 1 ] | |||
|
Location |
TSS200 (cg24396429) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.84E+00 | Statistic Test | p-value:1.20E-09; Z-score:1.03E+01 | ||
|
Methylation in Case |
7.21E-01 (Median) | Methylation in Control | 3.92E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
17 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC8A1 in breast cancer | [ 2 ] | |||
|
Location |
5'UTR (cg00610991) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.93E+00 | Statistic Test | p-value:9.67E-17; Z-score:3.48E+00 | ||
|
Methylation in Case |
3.51E-01 (Median) | Methylation in Control | 1.82E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC8A1 in breast cancer | [ 2 ] | |||
|
Location |
5'UTR (cg03256780) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.83E+00 | Statistic Test | p-value:1.34E-15; Z-score:-2.39E+00 | ||
|
Methylation in Case |
2.46E-01 (Median) | Methylation in Control | 4.50E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC8A1 in breast cancer | [ 2 ] | |||
|
Location |
5'UTR (cg15549810) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.19E+00 | Statistic Test | p-value:8.24E-12; Z-score:-2.45E+00 | ||
|
Methylation in Case |
7.10E-01 (Median) | Methylation in Control | 8.46E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC8A1 in breast cancer | [ 2 ] | |||
|
Location |
5'UTR (cg10633958) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.88E+00 | Statistic Test | p-value:8.26E-10; Z-score:2.78E+00 | ||
|
Methylation in Case |
1.24E-01 (Median) | Methylation in Control | 6.57E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC8A1 in breast cancer | [ 2 ] | |||
|
Location |
5'UTR (cg14411266) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.15E+00 | Statistic Test | p-value:1.69E-07; Z-score:1.27E+00 | ||
|
Methylation in Case |
9.18E-02 (Median) | Methylation in Control | 4.27E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC8A1 in breast cancer | [ 2 ] | |||
|
Location |
5'UTR (cg23601376) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.43E+00 | Statistic Test | p-value:2.53E-07; Z-score:1.48E+00 | ||
|
Methylation in Case |
8.23E-02 (Median) | Methylation in Control | 5.74E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC8A1 in breast cancer | [ 2 ] | |||
|
Location |
5'UTR (cg12748607) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.27E+00 | Statistic Test | p-value:2.67E-07; Z-score:1.19E+00 | ||
|
Methylation in Case |
7.58E-02 (Median) | Methylation in Control | 5.97E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC8A1 in breast cancer | [ 2 ] | |||
|
Location |
5'UTR (cg08240652) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:4.20E-06; Z-score:-1.71E+00 | ||
|
Methylation in Case |
8.30E-01 (Median) | Methylation in Control | 8.88E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC8A1 in breast cancer | [ 2 ] | |||
|
Location |
5'UTR (cg12742937) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:5.36E-06; Z-score:6.90E-01 | ||
|
Methylation in Case |
1.84E-01 (Median) | Methylation in Control | 1.61E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC8A1 in breast cancer | [ 2 ] | |||
|
Location |
5'UTR (cg03307465) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.33E+00 | Statistic Test | p-value:7.55E-06; Z-score:5.03E-01 | ||
|
Methylation in Case |
3.82E-02 (Median) | Methylation in Control | 2.87E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC8A1 in breast cancer | [ 2 ] | |||
|
Location |
5'UTR (cg09021626) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.23E+00 | Statistic Test | p-value:6.85E-05; Z-score:9.93E-01 | ||
|
Methylation in Case |
1.04E-01 (Median) | Methylation in Control | 8.40E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC8A1 in breast cancer | [ 2 ] | |||
|
Location |
5'UTR (cg20227806) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:2.89E-04; Z-score:7.06E-01 | ||
|
Methylation in Case |
8.03E-02 (Median) | Methylation in Control | 7.00E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC8A1 in breast cancer | [ 2 ] | |||
|
Location |
5'UTR (cg19918022) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:4.67E-04; Z-score:-6.17E-01 | ||
|
Methylation in Case |
8.47E-01 (Median) | Methylation in Control | 8.72E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC8A1 in breast cancer | [ 2 ] | |||
|
Location |
5'UTR (cg14686012) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:2.14E-02; Z-score:2.87E-01 | ||
|
Methylation in Case |
7.91E-02 (Median) | Methylation in Control | 7.25E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC8A1 in breast cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg02870829) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:6.57E-13; Z-score:-2.45E+00 | ||
|
Methylation in Case |
7.23E-01 (Median) | Methylation in Control | 8.27E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC8A1 in breast cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg02577240) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:8.05E-11; Z-score:-1.96E+00 | ||
|
Methylation in Case |
8.20E-01 (Median) | Methylation in Control | 8.81E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC8A1 in breast cancer | [ 2 ] | |||
|
Location |
TSS200 (cg24396429) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.39E+00 | Statistic Test | p-value:3.08E-18; Z-score:2.53E+00 | ||
|
Methylation in Case |
6.66E-01 (Median) | Methylation in Control | 4.78E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC8A1 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
5'UTR (cg14411266) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.52E+00 | Statistic Test | p-value:1.10E-04; Z-score:2.26E+00 | ||
|
Methylation in Case |
3.78E-02 (Median) | Methylation in Control | 2.48E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC8A1 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
5'UTR (cg09021626) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.18E+00 | Statistic Test | p-value:9.50E-03; Z-score:6.39E-01 | ||
|
Methylation in Case |
5.08E-02 (Median) | Methylation in Control | 4.30E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC8A1 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
5'UTR (cg12748607) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:1.20E-02; Z-score:5.18E-01 | ||
|
Methylation in Case |
2.55E-02 (Median) | Methylation in Control | 2.22E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC8A1 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
5'UTR (cg23601376) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:2.13E-02; Z-score:7.00E-01 | ||
|
Methylation in Case |
2.42E-02 (Median) | Methylation in Control | 2.03E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC8A1 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
5'UTR (cg14686012) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:2.14E-02; Z-score:3.72E-01 | ||
|
Methylation in Case |
3.63E-02 (Median) | Methylation in Control | 3.33E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC8A1 in clear cell renal cell carcinoma | [ 3 ] | |||
|
Location |
5'UTR (cg03307465) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:4.89E-02; Z-score:2.71E-01 | ||
|
Methylation in Case |
1.92E-02 (Median) | Methylation in Control | 1.81E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
16 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC8A1 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg14686012) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:3.11E+00 | Statistic Test | p-value:1.24E-18; Z-score:6.72E+00 | ||
|
Methylation in Case |
5.42E-01 (Median) | Methylation in Control | 1.74E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC8A1 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg03307465) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:4.24E+00 | Statistic Test | p-value:1.74E-18; Z-score:4.14E+00 | ||
|
Methylation in Case |
4.94E-01 (Median) | Methylation in Control | 1.17E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC8A1 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg09021626) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:3.02E+00 | Statistic Test | p-value:2.65E-17; Z-score:4.86E+00 | ||
|
Methylation in Case |
6.70E-01 (Median) | Methylation in Control | 2.22E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC8A1 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg20227806) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.30E+00 | Statistic Test | p-value:6.73E-15; Z-score:3.42E+00 | ||
|
Methylation in Case |
5.58E-01 (Median) | Methylation in Control | 2.42E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC8A1 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg14411266) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:3.03E+00 | Statistic Test | p-value:9.46E-14; Z-score:2.92E+00 | ||
|
Methylation in Case |
4.26E-01 (Median) | Methylation in Control | 1.40E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC8A1 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg12748607) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.96E+00 | Statistic Test | p-value:2.31E-12; Z-score:2.34E+00 | ||
|
Methylation in Case |
5.90E-01 (Median) | Methylation in Control | 3.01E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC8A1 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg12742937) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.57E+00 | Statistic Test | p-value:1.41E-10; Z-score:2.76E+00 | ||
|
Methylation in Case |
7.76E-01 (Median) | Methylation in Control | 4.95E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC8A1 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg08240652) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:8.93E-10; Z-score:-3.49E+00 | ||
|
Methylation in Case |
8.45E-01 (Median) | Methylation in Control | 9.27E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC8A1 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg15549810) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.19E+00 | Statistic Test | p-value:6.06E-09; Z-score:-2.64E+00 | ||
|
Methylation in Case |
6.44E-01 (Median) | Methylation in Control | 7.68E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC8A1 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg03256780) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.26E+00 | Statistic Test | p-value:2.67E-07; Z-score:-1.78E+00 | ||
|
Methylation in Case |
4.03E-01 (Median) | Methylation in Control | 5.06E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC8A1 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg19918022) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:2.30E-05; Z-score:-2.53E+00 | ||
|
Methylation in Case |
9.01E-01 (Median) | Methylation in Control | 9.24E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC8A1 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg10633958) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.33E+00 | Statistic Test | p-value:1.31E-04; Z-score:1.13E+00 | ||
|
Methylation in Case |
4.55E-01 (Median) | Methylation in Control | 3.42E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC8A1 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg00610991) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:5.05E-03; Z-score:9.53E-01 | ||
|
Methylation in Case |
6.46E-01 (Median) | Methylation in Control | 5.53E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC8A1 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg02870829) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:2.29E-05; Z-score:-9.79E-01 | ||
|
Methylation in Case |
7.53E-01 (Median) | Methylation in Control | 8.29E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC8A1 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg02577240) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:6.74E-05; Z-score:-8.00E-01 | ||
|
Methylation in Case |
7.58E-01 (Median) | Methylation in Control | 8.15E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC8A1 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS200 (cg13167631) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.24E+00 | Statistic Test | p-value:1.51E-04; Z-score:-1.11E+00 | ||
|
Methylation in Case |
6.41E-01 (Median) | Methylation in Control | 7.95E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
13 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC8A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
5'UTR (cg00610991) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.74E+00 | Statistic Test | p-value:1.19E-07; Z-score:-2.66E+00 | ||
|
Methylation in Case |
1.92E-01 (Median) | Methylation in Control | 3.33E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC8A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
5'UTR (cg03307465) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.51E+00 | Statistic Test | p-value:4.17E-06; Z-score:1.16E+00 | ||
|
Methylation in Case |
4.74E-02 (Median) | Methylation in Control | 3.14E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC8A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
5'UTR (cg23601376) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:6.32E-04; Z-score:-6.80E-01 | ||
|
Methylation in Case |
5.89E-02 (Median) | Methylation in Control | 7.99E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC8A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
5'UTR (cg09021626) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:1.26E-03; Z-score:5.37E-01 | ||
|
Methylation in Case |
1.53E-01 (Median) | Methylation in Control | 1.37E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC8A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
5'UTR (cg20227806) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.30E-03; Z-score:-3.52E-02 | ||
|
Methylation in Case |
9.28E-02 (Median) | Methylation in Control | 9.33E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC8A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
5'UTR (cg14686012) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.00E+00 | Statistic Test | p-value:1.46E-03; Z-score:1.67E-02 | ||
|
Methylation in Case |
8.49E-02 (Median) | Methylation in Control | 8.47E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC8A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
5'UTR (cg12748607) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:9.16E-03; Z-score:-3.75E-01 | ||
|
Methylation in Case |
7.14E-02 (Median) | Methylation in Control | 7.74E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC8A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
5'UTR (cg12742937) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.03E-02; Z-score:-2.44E-02 | ||
|
Methylation in Case |
1.60E-01 (Median) | Methylation in Control | 1.61E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC8A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg13913015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:3.56E+00 | Statistic Test | p-value:1.03E-09; Z-score:4.92E+00 | ||
|
Methylation in Case |
1.49E-01 (Median) | Methylation in Control | 4.18E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC8A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
1stExon (cg15668691) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.37E+00 | Statistic Test | p-value:1.55E-13; Z-score:-7.42E+00 | ||
|
Methylation in Case |
6.19E-01 (Median) | Methylation in Control | 8.50E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC8A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg18563642) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.51E+00 | Statistic Test | p-value:4.88E-16; Z-score:-3.28E+00 | ||
|
Methylation in Case |
3.69E-01 (Median) | Methylation in Control | 5.57E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC8A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg27382405) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.71E+00 | Statistic Test | p-value:2.63E-14; Z-score:2.25E+00 | ||
|
Methylation in Case |
5.16E-01 (Median) | Methylation in Control | 3.01E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC8A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg24369466) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:1.36E-11; Z-score:-4.53E+00 | ||
|
Methylation in Case |
8.15E-01 (Median) | Methylation in Control | 9.00E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC8A1 in lung adenocarcinoma | [ 6 ] | |||
|
Location |
5'UTR (cg03307465) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:3.99E+00 | Statistic Test | p-value:7.71E-03; Z-score:1.27E+01 | ||
|
Methylation in Case |
1.75E-01 (Median) | Methylation in Control | 4.38E-02 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC8A1 in lung adenocarcinoma | [ 6 ] | |||
|
Location |
5'UTR (cg10633958) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.40E+00 | Statistic Test | p-value:1.33E-02; Z-score:2.42E+00 | ||
|
Methylation in Case |
1.67E-01 (Median) | Methylation in Control | 1.19E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC8A1 in lung adenocarcinoma | [ 6 ] | |||
|
Location |
5'UTR (cg14411266) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.14E+00 | Statistic Test | p-value:1.69E-02; Z-score:5.21E+00 | ||
|
Methylation in Case |
1.82E-01 (Median) | Methylation in Control | 8.50E-02 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC8A1 in lung adenocarcinoma | [ 6 ] | |||
|
Location |
5'UTR (cg09021626) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.24E+00 | Statistic Test | p-value:2.91E-02; Z-score:1.70E+00 | ||
|
Methylation in Case |
1.64E-01 (Median) | Methylation in Control | 1.33E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC8A1 in lung adenocarcinoma | [ 6 ] | |||
|
Location |
5'UTR (cg19918022) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:4.77E-02; Z-score:-1.49E+00 | ||
|
Methylation in Case |
8.67E-01 (Median) | Methylation in Control | 8.87E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC8A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
5'UTR (cg08240652) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:8.42E-05; Z-score:-6.27E-01 | ||
|
Methylation in Case |
9.05E-01 (Median) | Methylation in Control | 9.21E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC8A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
5'UTR (cg03256780) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:1.13E-02; Z-score:-7.04E-01 | ||
|
Methylation in Case |
3.16E-01 (Median) | Methylation in Control | 3.58E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC8A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
5'UTR (cg19918022) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.39E-02; Z-score:-2.89E-01 | ||
|
Methylation in Case |
9.25E-01 (Median) | Methylation in Control | 9.30E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC8A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg02577240) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:9.95E-03; Z-score:-3.53E-01 | ||
|
Methylation in Case |
9.31E-01 (Median) | Methylation in Control | 9.36E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC8A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg02870829) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.32E-02; Z-score:-2.72E-01 | ||
|
Methylation in Case |
8.99E-01 (Median) | Methylation in Control | 9.06E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC8A1 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
TSS200 (cg24396429) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:1.24E-06; Z-score:1.08E+00 | ||
|
Methylation in Case |
7.57E-01 (Median) | Methylation in Control | 6.92E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Pancretic ductal adenocarcinoma |
10 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC8A1 in pancretic ductal adenocarcinoma | [ 8 ] | |||
|
Location |
TSS1500 (cg27485921) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.27E+00 | Statistic Test | p-value:8.24E-10; Z-score:-1.87E+00 | ||
|
Methylation in Case |
4.60E-01 (Median) | Methylation in Control | 5.86E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC8A1 in pancretic ductal adenocarcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg27569822) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:3.16E-07; Z-score:1.02E+00 | ||
|
Methylation in Case |
6.87E-02 (Median) | Methylation in Control | 5.85E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC8A1 in pancretic ductal adenocarcinoma | [ 8 ] | |||
|
Location |
Body (cg11666857) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:3.24E-22; Z-score:-3.38E+00 | ||
|
Methylation in Case |
6.60E-01 (Median) | Methylation in Control | 8.03E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC8A1 in pancretic ductal adenocarcinoma | [ 8 ] | |||
|
Location |
Body (cg10245915) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.88E+00 | Statistic Test | p-value:2.26E-19; Z-score:2.96E+00 | ||
|
Methylation in Case |
3.17E-01 (Median) | Methylation in Control | 1.69E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC8A1 in pancretic ductal adenocarcinoma | [ 8 ] | |||
|
Location |
Body (cg24454829) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.34E+00 | Statistic Test | p-value:8.62E-19; Z-score:2.89E+00 | ||
|
Methylation in Case |
4.36E-01 (Median) | Methylation in Control | 3.26E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC8A1 in pancretic ductal adenocarcinoma | [ 8 ] | |||
|
Location |
Body (cg05978154) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.23E+00 | Statistic Test | p-value:1.22E-10; Z-score:-1.79E+00 | ||
|
Methylation in Case |
6.89E-01 (Median) | Methylation in Control | 8.48E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC8A1 in pancretic ductal adenocarcinoma | [ 8 ] | |||
|
Location |
Body (cg06094523) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.33E+00 | Statistic Test | p-value:3.55E-10; Z-score:-1.47E+00 | ||
|
Methylation in Case |
3.97E-01 (Median) | Methylation in Control | 5.29E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC8A1 in pancretic ductal adenocarcinoma | [ 8 ] | |||
|
Location |
Body (cg05169099) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:6.71E-06; Z-score:1.25E+00 | ||
|
Methylation in Case |
7.99E-01 (Median) | Methylation in Control | 7.07E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC8A1 in pancretic ductal adenocarcinoma | [ 8 ] | |||
|
Location |
Body (cg00718541) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:1.44E-05; Z-score:8.11E-01 | ||
|
Methylation in Case |
9.06E-01 (Median) | Methylation in Control | 8.79E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC8A1 in pancretic ductal adenocarcinoma | [ 8 ] | |||
|
Location |
Body (cg09691393) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:7.04E-03; Z-score:8.12E-01 | ||
|
Methylation in Case |
8.58E-01 (Median) | Methylation in Control | 8.14E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC8A1 in panic disorder | [ 9 ] | |||
|
Location |
TSS1500 (cg02870829) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-3.18E+00 | Statistic Test | p-value:1.99E-04; Z-score:-5.91E-01 | ||
|
Methylation in Case |
1.20E-01 (Median) | Methylation in Control | 3.83E-01 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colon cancer |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC8A1 in colon adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg07546508) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:3.30E-05; Z-score:-1.88E+00 | ||
|
Methylation in Case |
5.76E-01 (Median) | Methylation in Control | 6.57E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC8A1 in colon adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg06728970) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:1.44E-03; Z-score:-1.22E+00 | ||
|
Methylation in Case |
4.95E-01 (Median) | Methylation in Control | 5.64E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC8A1 in colon adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg24948829) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.83E-03; Z-score:-1.57E+00 | ||
|
Methylation in Case |
7.46E-01 (Median) | Methylation in Control | 7.76E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC8A1 in colon adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg15647725) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:3.21E-03; Z-score:9.76E-01 | ||
|
Methylation in Case |
6.22E-01 (Median) | Methylation in Control | 5.94E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC8A1 in colon adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg19272348) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:3.22E-03; Z-score:-1.69E+00 | ||
|
Methylation in Case |
4.91E-01 (Median) | Methylation in Control | 5.78E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC8A1 in prostate cancer | [ 11 ] | |||
|
Location |
Body (cg04204452) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.29E+00 | Statistic Test | p-value:1.58E-03; Z-score:3.92E+00 | ||
|
Methylation in Case |
8.49E-01 (Median) | Methylation in Control | 6.61E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC8A1 in prostate cancer | [ 11 ] | |||
|
Location |
Body (cg23707719) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.35E+00 | Statistic Test | p-value:5.35E-03; Z-score:8.83E+00 | ||
|
Methylation in Case |
8.48E-01 (Median) | Methylation in Control | 6.27E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC8A1 in prostate cancer | [ 11 ] | |||
|
Location |
Body (cg00704369) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:8.12E-03; Z-score:2.85E+00 | ||
|
Methylation in Case |
9.27E-01 (Median) | Methylation in Control | 8.47E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC8A1 in prostate cancer | [ 11 ] | |||
|
Location |
Body (cg02057782) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.58E+00 | Statistic Test | p-value:1.33E-02; Z-score:1.11E+01 | ||
|
Methylation in Case |
6.44E-01 (Median) | Methylation in Control | 4.08E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC8A1 in prostate cancer | [ 11 ] | |||
|
Location |
Body (cg24724513) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.78E+00 | Statistic Test | p-value:1.72E-02; Z-score:-7.82E+00 | ||
|
Methylation in Case |
4.37E-01 (Median) | Methylation in Control | 7.80E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC8A1 in prostate cancer | [ 11 ] | |||
|
Location |
3'UTR (cg08203794) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:6.52E-03; Z-score:2.93E+00 | ||
|
Methylation in Case |
8.91E-01 (Median) | Methylation in Control | 8.50E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC8A1 in prostate cancer | [ 11 ] | |||
|
Location |
3'UTR (cg05174446) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:2.24E-02; Z-score:2.96E+00 | ||
|
Methylation in Case |
8.81E-01 (Median) | Methylation in Control | 8.33E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC8A1 in prostate cancer | [ 11 ] | |||
|
Location |
3'UTR (cg08241225) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:2.95E-02; Z-score:1.52E+00 | ||
|
Methylation in Case |
9.03E-01 (Median) | Methylation in Control | 8.32E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
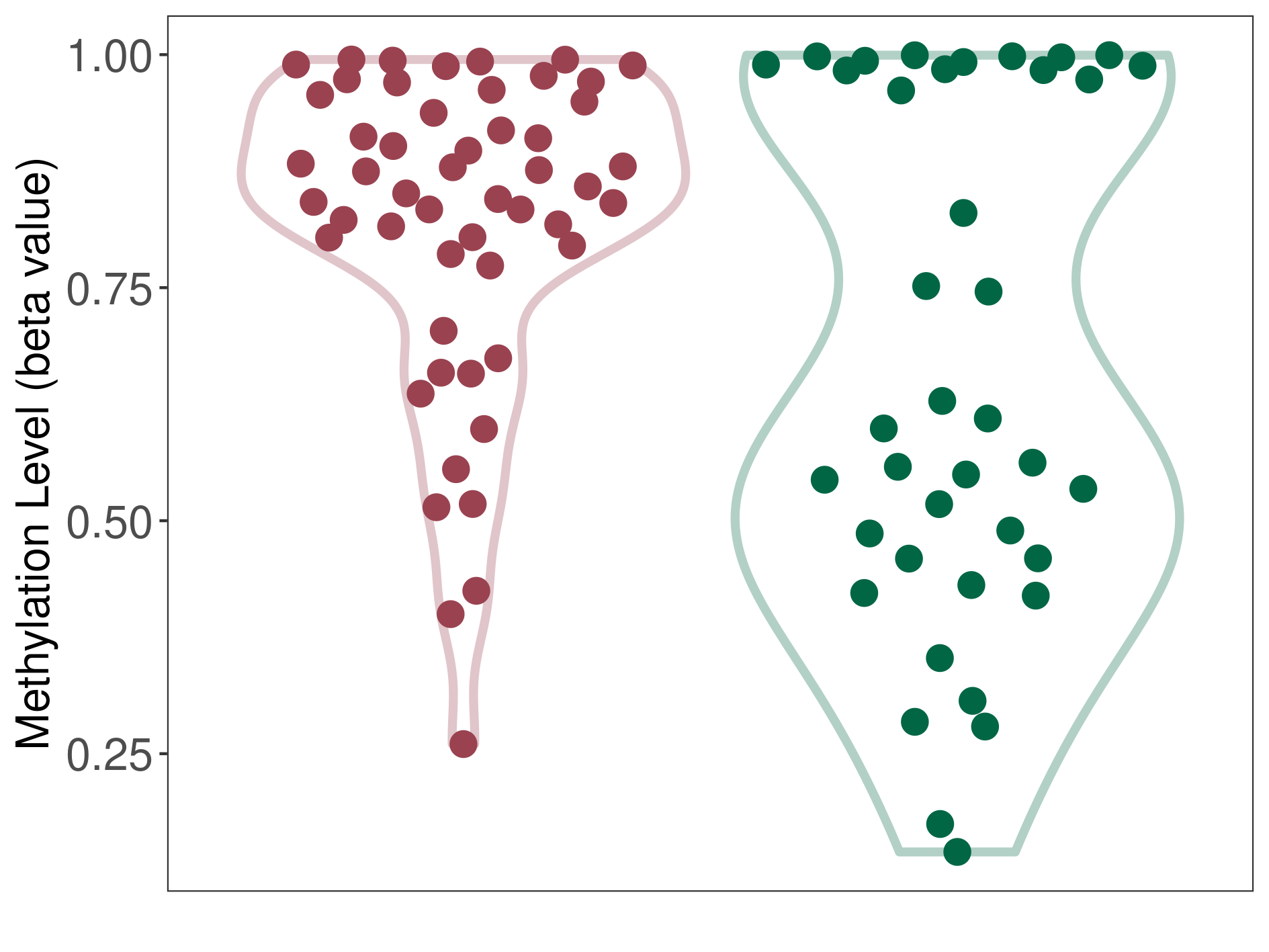
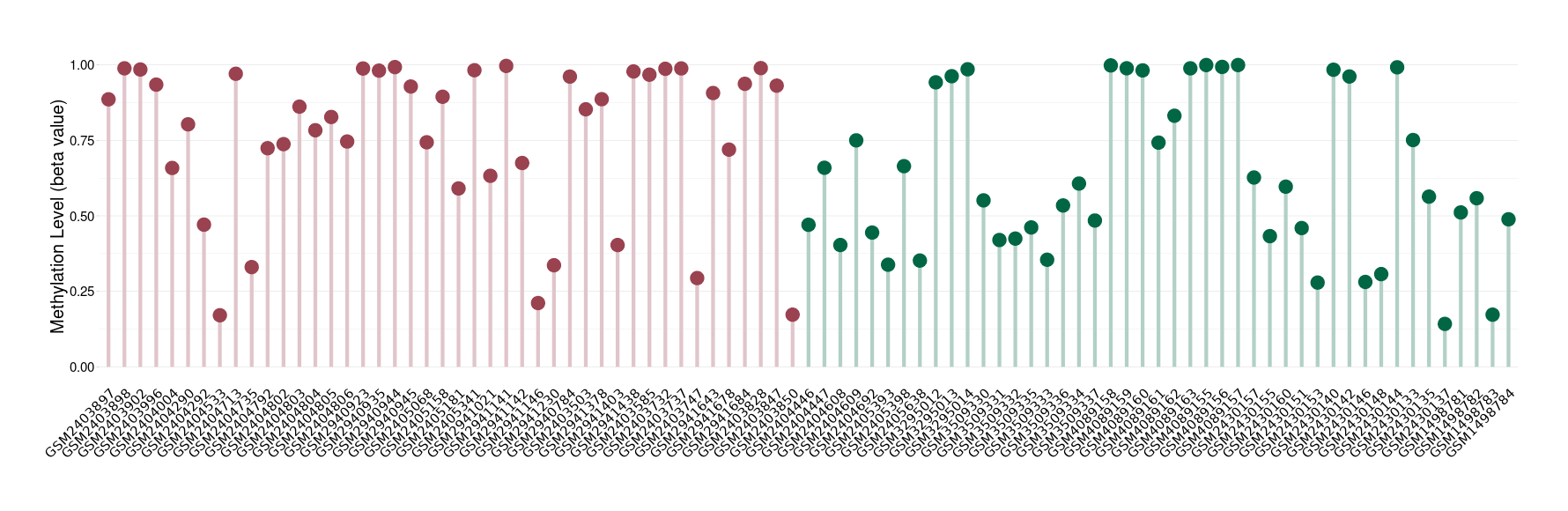
Multilayered rosettes embryonal tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypermethylation of SLC8A1 in multilayered rosettes embryonal tumour than that in healthy individual | ||||
Studied Phenotype |
Multilayered rosettes embryonal tumour [ICD-11:2A00.1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.004129274; Fold-change:0.255889435; Z-score:0.916987402 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
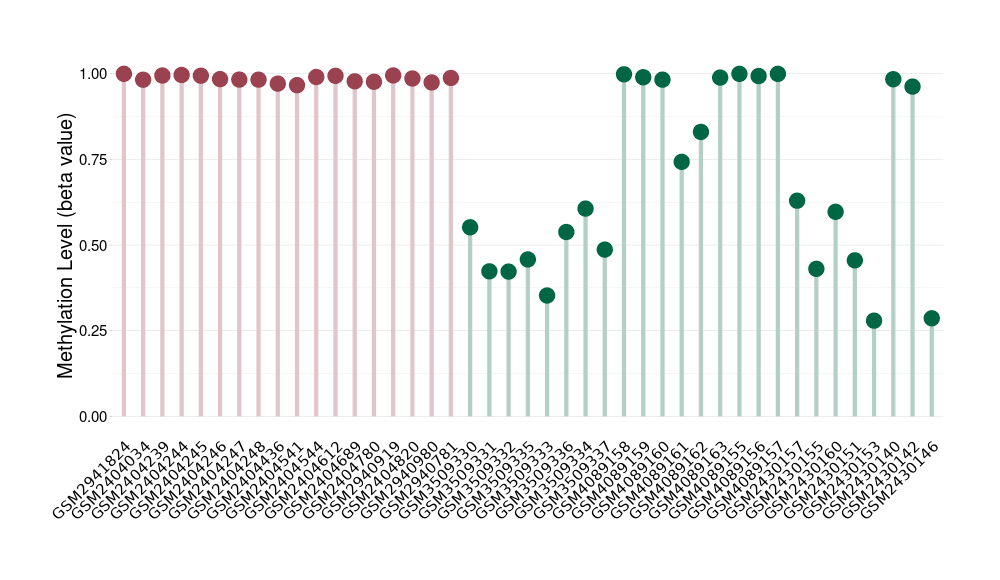
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
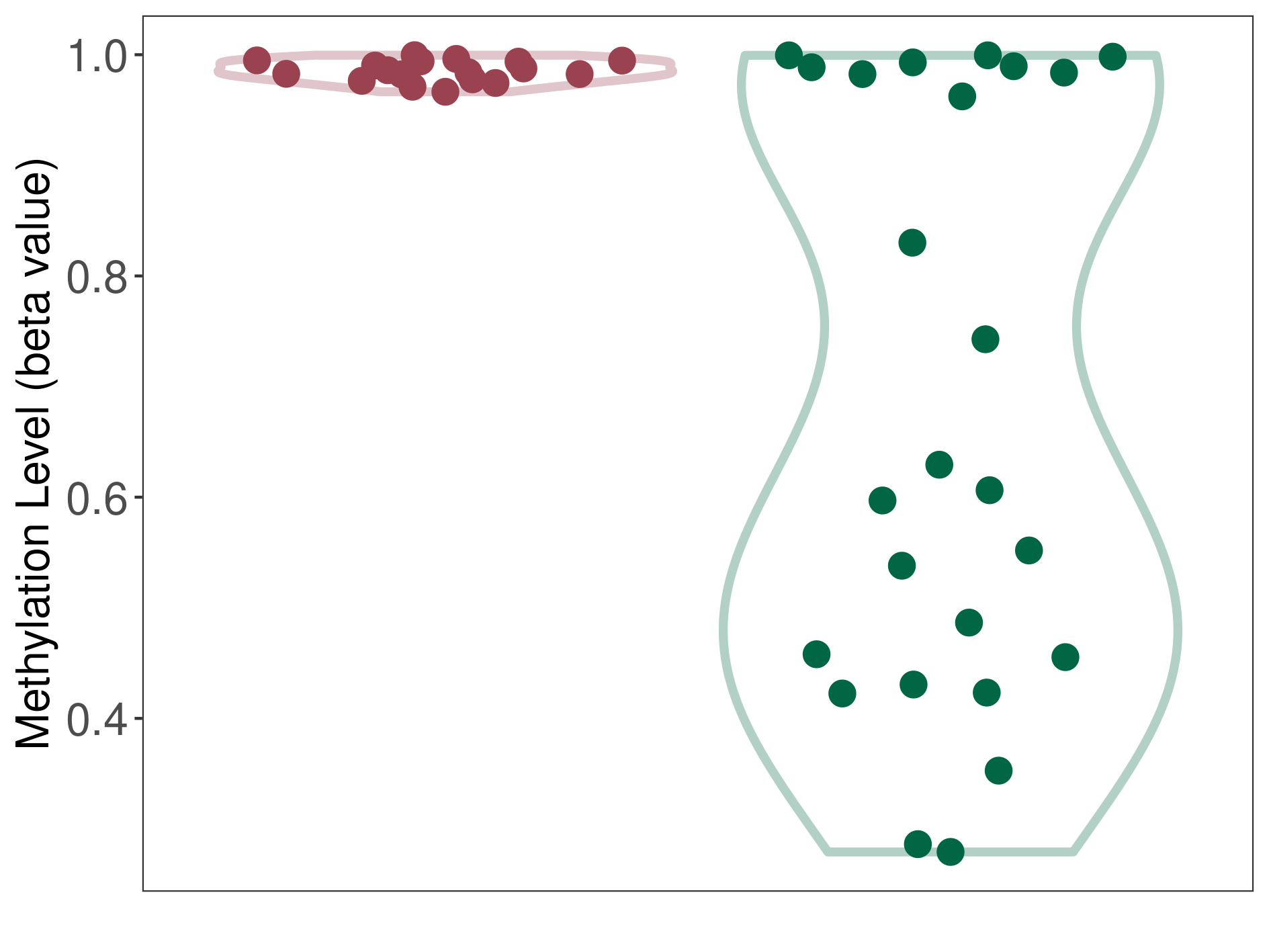
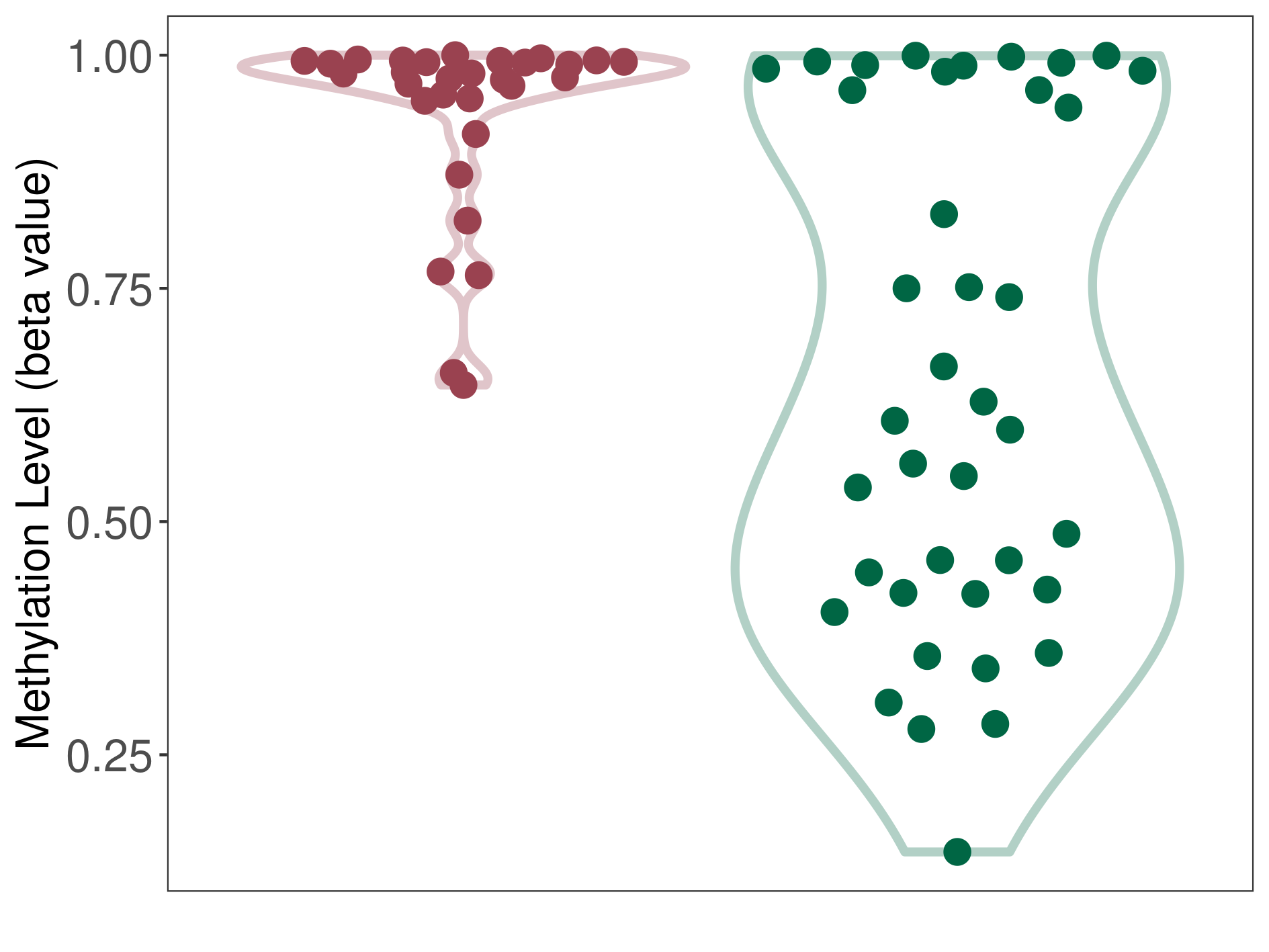
Melanocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC8A1 in melanocytoma than that in healthy individual | ||||
Studied Phenotype |
Melanocytoma [ICD-11:2F36.2] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:6.15E-06; Fold-change:0.378751745; Z-score:1.427249903 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
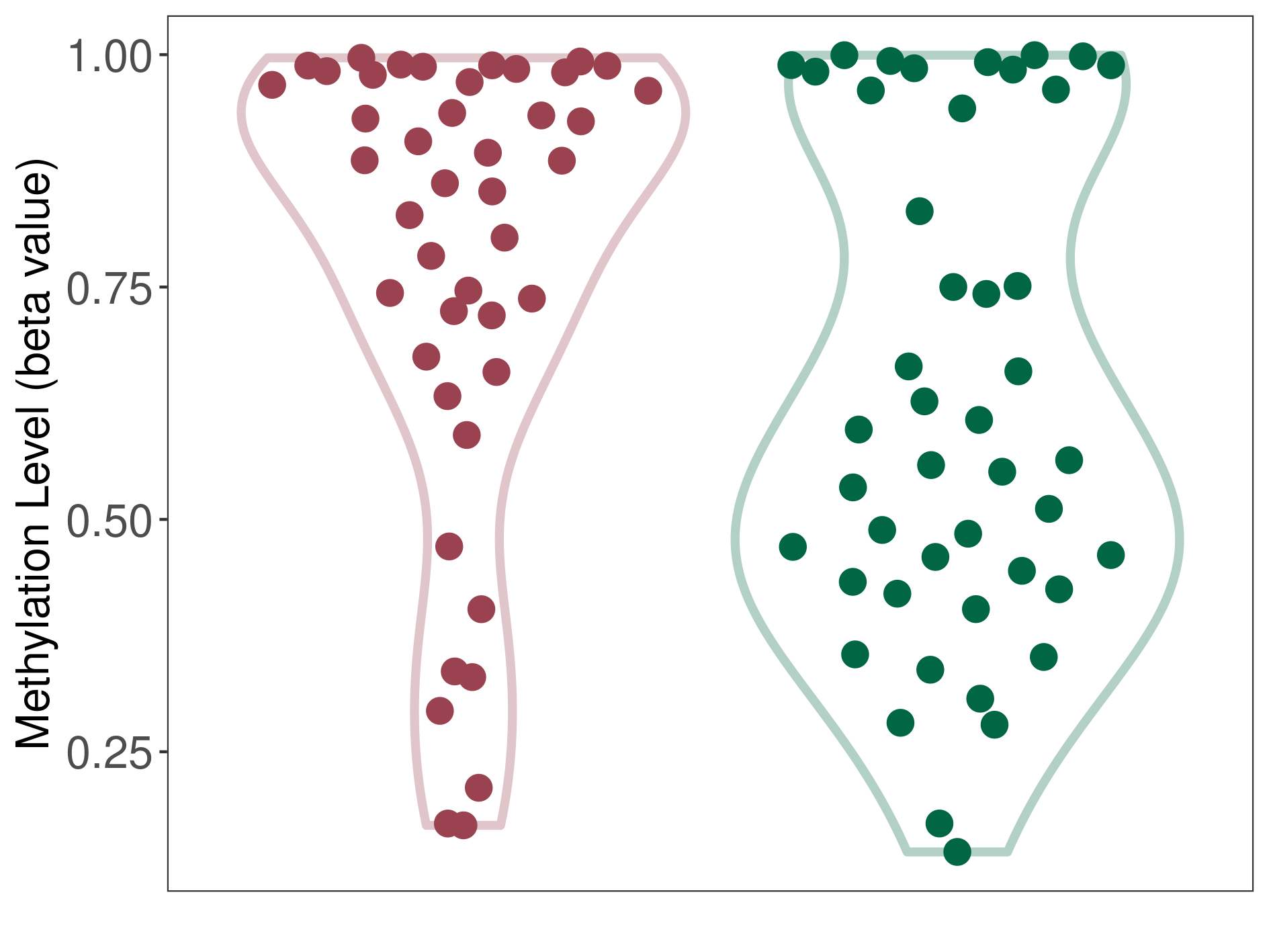
Myxopapillary ependymoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC8A1 in myxopapillary ependymoma than that in healthy individual | ||||
Studied Phenotype |
Myxopapillary ependymoma [ICD-11:2A00.5] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.015285926; Fold-change:0.310090744; Z-score:1.16535805 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Peripheral neuroectodermal tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC8A1 in peripheral neuroectodermal tumour than that in healthy individual | ||||
Studied Phenotype |
Peripheral neuroectodermal tumour [ICD-11:2B52] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:3.06E-07; Fold-change:0.370115587; Z-score:1.361646643 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
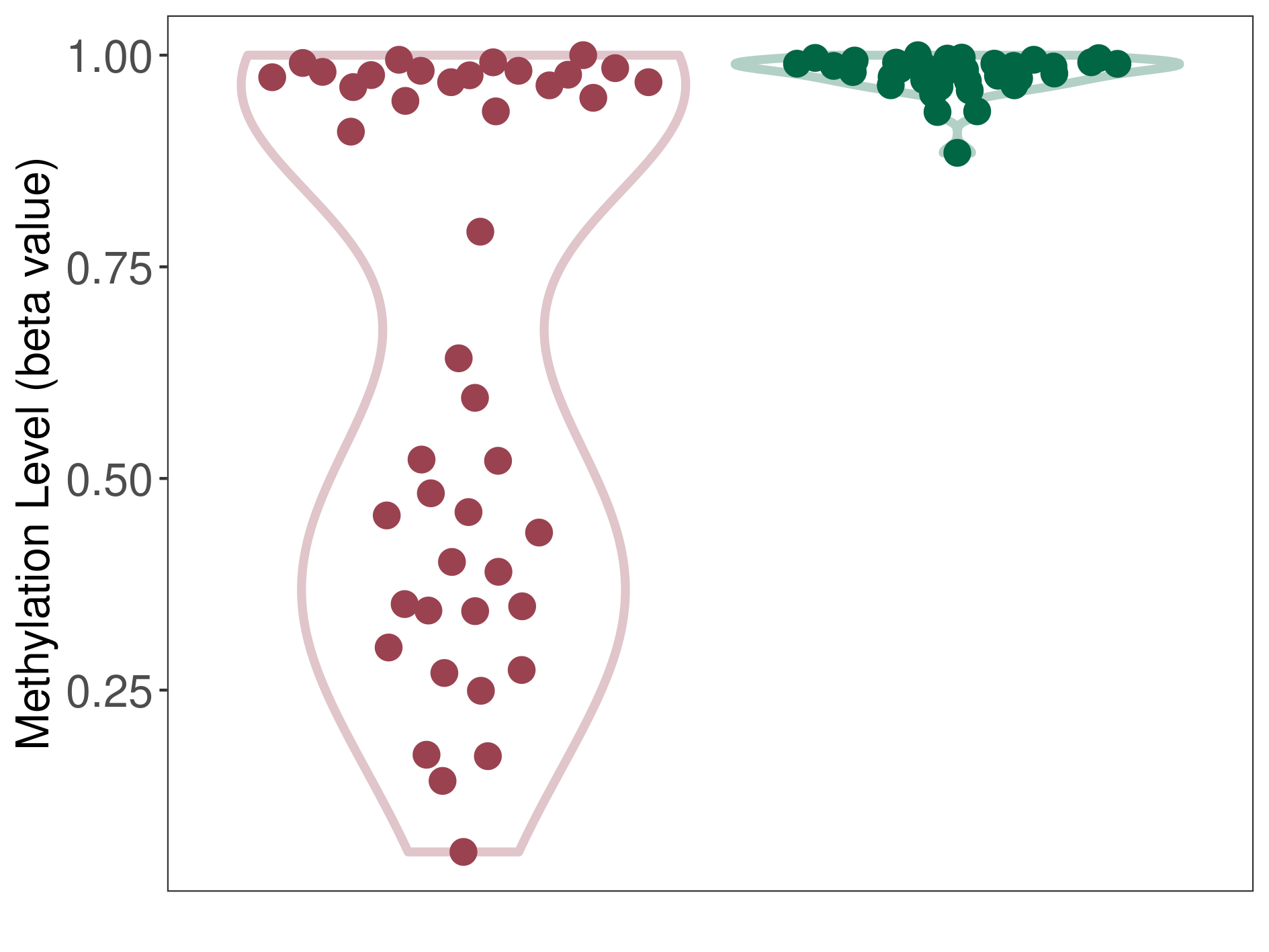
Craniopharyngioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC8A1 in craniopharyngioma than that in healthy individual | ||||
Studied Phenotype |
Craniopharyngioma [ICD-11:2F9A] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:6.99E-08; Fold-change:-0.341285526; Z-score:-14.64442798 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
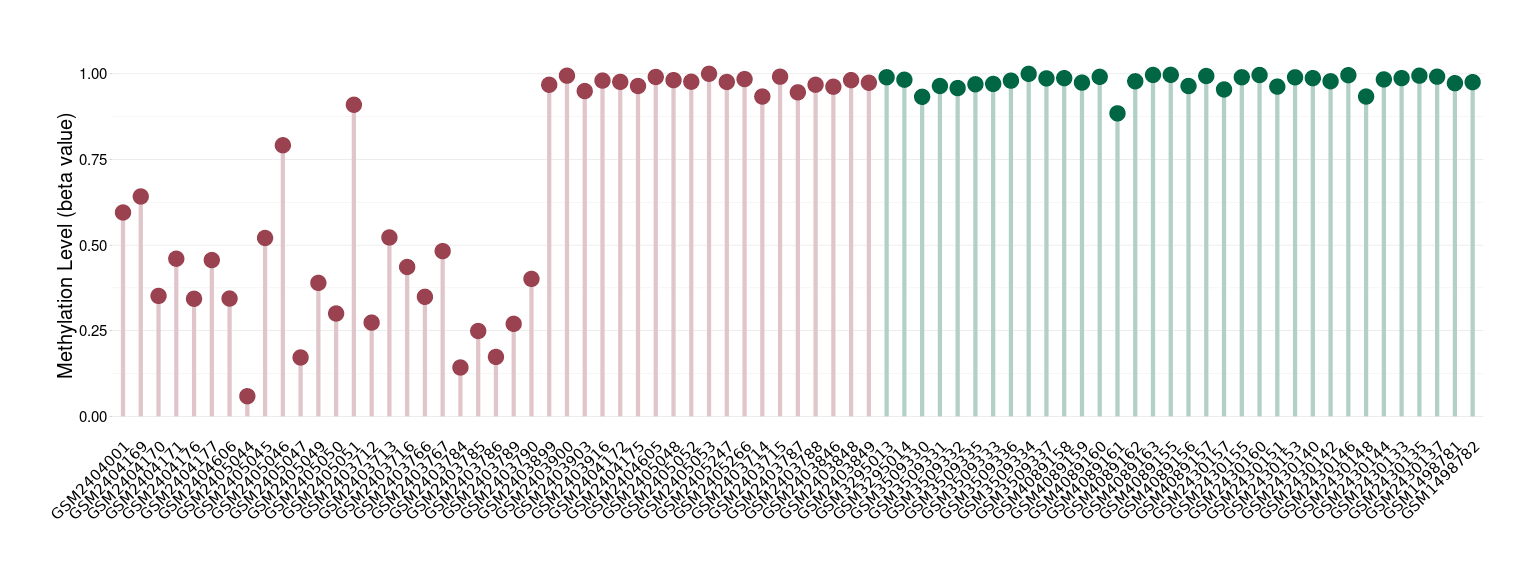
Posterior fossa ependymoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC8A1 in posterior fossa ependymoma than that in healthy individual | ||||
Studied Phenotype |
Posterior fossa ependymoma [ICD-11:2D50.2] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:2.14E-37; Fold-change:-0.43152921; Z-score:-1.970444783 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
microRNA |
|||||
|
Unclear Phenotype |
42 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
miR-1305 directly targets SLC8A1 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-1305 | miRNA Mature ID | miR-1305 | ||
|
miRNA Sequence |
UUUUCAACUCUAAUGGGAGAGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon2 |
miR-146a directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-146a | miRNA Mature ID | miR-146a-3p | ||
|
miRNA Sequence |
CCUCUGAAAUUCAGUUCUUCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon3 |
miR-1825 directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1825 | miRNA Mature ID | miR-1825 | ||
|
miRNA Sequence |
UCCAGUGCCCUCCUCUCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon4 |
miR-190a directly targets SLC8A1 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP//HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-190a | miRNA Mature ID | miR-190a-3p | ||
|
miRNA Sequence |
CUAUAUAUCAAACAUAUUCCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon5 |
miR-199a directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-199a | miRNA Mature ID | miR-199a-5p | ||
|
miRNA Sequence |
CCCAGUGUUCAGACUACCUGUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon6 |
miR-199b directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-199b | miRNA Mature ID | miR-199b-5p | ||
|
miRNA Sequence |
CCCAGUGUUUAGACUAUCUGUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon7 |
miR-3135a directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3135a | miRNA Mature ID | miR-3135a | ||
|
miRNA Sequence |
UGCCUAGGCUGAGACUGCAGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon8 |
miR-3148 directly targets SLC8A1 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3148 | miRNA Mature ID | miR-3148 | ||
|
miRNA Sequence |
UGGAAAAAACUGGUGUGUGCUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon9 |
miR-3150b directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3150b | miRNA Mature ID | miR-3150b-3p | ||
|
miRNA Sequence |
UGAGGAGAUCGUCGAGGUUGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon10 |
miR-3161 directly targets SLC8A1 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3161 | miRNA Mature ID | miR-3161 | ||
|
miRNA Sequence |
CUGAUAAGAACAGAGGCCCAGAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon11 |
miR-3606 directly targets SLC8A1 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3606 | miRNA Mature ID | miR-3606-3p | ||
|
miRNA Sequence |
AAAAUUUCUUUCACUACUUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon12 |
miR-3622a directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3622a | miRNA Mature ID | miR-3622a-5p | ||
|
miRNA Sequence |
CAGGCACGGGAGCUCAGGUGAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon13 |
miR-3924 directly targets SLC8A1 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP//HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3924 | miRNA Mature ID | miR-3924 | ||
|
miRNA Sequence |
AUAUGUAUAUGUGACUGCUACU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon14 |
miR-3978 directly targets SLC8A1 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3978 | miRNA Mature ID | miR-3978 | ||
|
miRNA Sequence |
GUGGAAAGCAUGCAUCCAGGGUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon15 |
miR-410 directly targets SLC8A1 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP//HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-410 | miRNA Mature ID | miR-410-3p | ||
|
miRNA Sequence |
AAUAUAACACAGAUGGCCUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon16 |
miR-4282 directly targets SLC8A1 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4282 | miRNA Mature ID | miR-4282 | ||
|
miRNA Sequence |
UAAAAUUUGCAUCCAGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon17 |
miR-4423 directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4423 | miRNA Mature ID | miR-4423-3p | ||
|
miRNA Sequence |
AUAGGCACCAAAAAGCAACAA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon18 |
miR-4668 directly targets SLC8A1 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4668 | miRNA Mature ID | miR-4668-3p | ||
|
miRNA Sequence |
GAAAAUCCUUUUUGUUUUUCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon19 |
miR-4683 directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4683 | miRNA Mature ID | miR-4683 | ||
|
miRNA Sequence |
UGGAGAUCCAGUGCUCGCCCGAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon20 |
miR-4699 directly targets SLC8A1 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4699 | miRNA Mature ID | miR-4699-3p | ||
|
miRNA Sequence |
AAUUUACUCUGCAAUCUUCUCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon21 |
miR-4766 directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4766 | miRNA Mature ID | miR-4766-5p | ||
|
miRNA Sequence |
UCUGAAAGAGCAGUUGGUGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon22 |
miR-4775 directly targets SLC8A1 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4775 | miRNA Mature ID | miR-4775 | ||
|
miRNA Sequence |
UUAAUUUUUUGUUUCGGUCACU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon23 |
miR-4784 directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4784 | miRNA Mature ID | miR-4784 | ||
|
miRNA Sequence |
UGAGGAGAUGCUGGGACUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon24 |
miR-494 directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-494 | miRNA Mature ID | miR-494-3p | ||
|
miRNA Sequence |
UGAAACAUACACGGGAAACCUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon25 |
miR-500a directly targets SLC8A1 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-500a | miRNA Mature ID | miR-500a-5p | ||
|
miRNA Sequence |
UAAUCCUUGCUACCUGGGUGAGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon26 |
miR-501 directly targets SLC8A1 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-501 | miRNA Mature ID | miR-501-5p | ||
|
miRNA Sequence |
AAUCCUUUGUCCCUGGGUGAGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon27 |
miR-5011 directly targets SLC8A1 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP//HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5011 | miRNA Mature ID | miR-5011-5p | ||
|
miRNA Sequence |
UAUAUAUACAGCCAUGCACUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon28 |
miR-507 directly targets SLC8A1 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-507 | miRNA Mature ID | miR-507 | ||
|
miRNA Sequence |
UUUUGCACCUUUUGGAGUGAA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon29 |
miR-512 directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-512 | miRNA Mature ID | miR-512-3p | ||
|
miRNA Sequence |
AAGUGCUGUCAUAGCUGAGGUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon30 |
miR-513a directly targets SLC8A1 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-513a | miRNA Mature ID | miR-513a-3p | ||
|
miRNA Sequence |
UAAAUUUCACCUUUCUGAGAAGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon31 |
miR-513c directly targets SLC8A1 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-513c | miRNA Mature ID | miR-513c-3p | ||
|
miRNA Sequence |
UAAAUUUCACCUUUCUGAGAAGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon32 |
miR-557 directly targets SLC8A1 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-557 | miRNA Mature ID | miR-557 | ||
|
miRNA Sequence |
GUUUGCACGGGUGGGCCUUGUCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon33 |
miR-5582 directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-5582 | miRNA Mature ID | miR-5582-5p | ||
|
miRNA Sequence |
UAGGCACACUUAAAGUUAUAGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon34 |
miR-5693 directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-5693 | miRNA Mature ID | miR-5693 | ||
|
miRNA Sequence |
GCAGUGGCUCUGAAAUGAACUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon35 |
miR-590 directly targets SLC8A1 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP//HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-590 | miRNA Mature ID | miR-590-3p | ||
|
miRNA Sequence |
UAAUUUUAUGUAUAAGCUAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon36 |
miR-599 directly targets SLC8A1 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-599 | miRNA Mature ID | miR-599 | ||
|
miRNA Sequence |
GUUGUGUCAGUUUAUCAAAC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon37 |
miR-6124 directly targets SLC8A1 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6124 | miRNA Mature ID | miR-6124 | ||
|
miRNA Sequence |
GGGAAAAGGAAGGGGGAGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon38 |
miR-6822 directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6822 | miRNA Mature ID | miR-6822-3p | ||
|
miRNA Sequence |
AGGCUCUAACUGGCUUUCCCUGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon39 |
miR-6847 directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6847 | miRNA Mature ID | miR-6847-5p | ||
|
miRNA Sequence |
ACAGAGGACAGUGGAGUGUGAGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon40 |
miR-6888 directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6888 | miRNA Mature ID | miR-6888-5p | ||
|
miRNA Sequence |
AAGGAGAUGCUCAGGCAGAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon41 |
miR-760 directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-760 | miRNA Mature ID | miR-760 | ||
|
miRNA Sequence |
CGGCUCUGGGUCUGUGGGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon42 |
miR-8057 directly targets SLC8A1 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-8057 | miRNA Mature ID | miR-8057 | ||
|
miRNA Sequence |
GUGGCUCUGUAGUAAGAUGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
