Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0464 Transporter Info | ||||
| Gene Name | SLC7A11 | ||||
| Transporter Name | Cystine/glutamate transporter | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Pancretic ductal adenocarcinoma |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC7A11 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg10265016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.29E+00 | Statistic Test | p-value:1.02E-06; Z-score:1.35E+00 | ||
|
Methylation in Case |
5.69E-01 (Median) | Methylation in Control | 4.41E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC7A11 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg18935813) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.80E+00 | Statistic Test | p-value:5.22E-18; Z-score:1.56E+00 | ||
|
Methylation in Case |
7.22E-02 (Median) | Methylation in Control | 4.02E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC7A11 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg07354253) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:1.98E-07; Z-score:-1.35E+00 | ||
|
Methylation in Case |
1.23E-01 (Median) | Methylation in Control | 1.51E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC7A11 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
1stExon (cg25437385) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.35E+00 | Statistic Test | p-value:5.50E-07; Z-score:4.92E-01 | ||
|
Methylation in Case |
7.68E-02 (Median) | Methylation in Control | 5.68E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC7A11 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg03731131) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.42E+00 | Statistic Test | p-value:2.18E-07; Z-score:1.55E+00 | ||
|
Methylation in Case |
6.38E-01 (Median) | Methylation in Control | 4.49E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC7A11 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg24733435) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:4.47E-03; Z-score:7.17E-01 | ||
|
Methylation in Case |
7.93E-01 (Median) | Methylation in Control | 7.73E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC7A11 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg04433051) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.39E+00 | Statistic Test | p-value:6.56E-03; Z-score:-9.30E-01 | ||
|
Methylation in Case |
3.47E-01 (Median) | Methylation in Control | 4.84E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC7A11 in breast cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg00361146) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:1.55E-02; Z-score:4.19E-01 | ||
|
Methylation in Case |
4.76E-02 (Median) | Methylation in Control | 4.06E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC7A11 in breast cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg02102889) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.04E-02; Z-score:-4.66E-01 | ||
|
Methylation in Case |
7.06E-01 (Median) | Methylation in Control | 7.42E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC7A11 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg21877274) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:4.72E-12; Z-score:-2.66E+00 | ||
|
Methylation in Case |
8.24E-01 (Median) | Methylation in Control | 8.85E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC7A11 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg24869834) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:8.41E-10; Z-score:-1.78E+00 | ||
|
Methylation in Case |
7.52E-01 (Median) | Methylation in Control | 8.47E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC7A11 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg02734904) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:2.88E-07; Z-score:-1.42E+00 | ||
|
Methylation in Case |
8.59E-01 (Median) | Methylation in Control | 8.95E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC7A11 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg01309945) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:5.23E-05; Z-score:-1.22E+00 | ||
|
Methylation in Case |
8.43E-01 (Median) | Methylation in Control | 8.67E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC7A11 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg04487857) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:5.55E-05; Z-score:1.88E+00 | ||
|
Methylation in Case |
6.12E-01 (Median) | Methylation in Control | 5.06E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC7A11 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg06206831) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:8.72E-05; Z-score:-1.09E+00 | ||
|
Methylation in Case |
8.75E-01 (Median) | Methylation in Control | 8.93E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC7A11 in breast cancer | [ 2 ] | |||
|
Location |
Body (cg15867963) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:1.25E-04; Z-score:1.35E+00 | ||
|
Methylation in Case |
6.73E-01 (Median) | Methylation in Control | 5.95E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC7A11 in colorectal cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg02102889) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.20E+00 | Statistic Test | p-value:4.78E-08; Z-score:-2.02E+00 | ||
|
Methylation in Case |
7.50E-01 (Median) | Methylation in Control | 9.03E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC7A11 in colorectal cancer | [ 3 ] | |||
|
Location |
Body (cg15867963) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.37E+00 | Statistic Test | p-value:2.01E-07; Z-score:1.77E+00 | ||
|
Methylation in Case |
7.51E-01 (Median) | Methylation in Control | 5.47E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC7A11 in colorectal cancer | [ 3 ] | |||
|
Location |
Body (cg01309945) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:3.45E-07; Z-score:-1.78E+00 | ||
|
Methylation in Case |
8.72E-01 (Median) | Methylation in Control | 9.02E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC7A11 in colorectal cancer | [ 3 ] | |||
|
Location |
Body (cg04487857) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.42E+00 | Statistic Test | p-value:1.75E-05; Z-score:1.54E+00 | ||
|
Methylation in Case |
6.75E-01 (Median) | Methylation in Control | 4.76E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC7A11 in colorectal cancer | [ 3 ] | |||
|
Location |
Body (cg15441006) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:3.09E-02; Z-score:-5.70E-01 | ||
|
Methylation in Case |
7.73E-01 (Median) | Methylation in Control | 7.97E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
17 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC7A11 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
TSS1500 (cg02102889) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:5.81E-08; Z-score:-8.95E-01 | ||
|
Methylation in Case |
7.03E-01 (Median) | Methylation in Control | 7.76E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC7A11 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
1stExon (cg16399511) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.59E+00 | Statistic Test | p-value:2.78E-23; Z-score:-4.76E+00 | ||
|
Methylation in Case |
4.45E-01 (Median) | Methylation in Control | 7.08E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC7A11 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
Body (cg21460403) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.01E+00 | Statistic Test | p-value:4.49E-22; Z-score:-5.12E+00 | ||
|
Methylation in Case |
3.38E-01 (Median) | Methylation in Control | 6.79E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC7A11 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
Body (cg07845352) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.72E+00 | Statistic Test | p-value:3.05E-17; Z-score:-2.86E+00 | ||
|
Methylation in Case |
3.75E-01 (Median) | Methylation in Control | 6.46E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC7A11 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
Body (cg20141652) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:2.65E-12; Z-score:1.87E+00 | ||
|
Methylation in Case |
5.21E-01 (Median) | Methylation in Control | 4.30E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC7A11 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
Body (cg19063061) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.25E+00 | Statistic Test | p-value:5.72E-11; Z-score:3.86E+00 | ||
|
Methylation in Case |
4.57E-01 (Median) | Methylation in Control | 2.03E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC7A11 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
Body (cg06690548) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.25E+00 | Statistic Test | p-value:9.71E-09; Z-score:-1.89E+00 | ||
|
Methylation in Case |
5.53E-01 (Median) | Methylation in Control | 6.94E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC7A11 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
Body (cg02734904) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:4.02E-08; Z-score:-2.11E+00 | ||
|
Methylation in Case |
7.36E-01 (Median) | Methylation in Control | 8.16E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC7A11 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
Body (cg21877274) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:9.67E-08; Z-score:-2.65E+00 | ||
|
Methylation in Case |
7.19E-01 (Median) | Methylation in Control | 8.14E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC7A11 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
Body (cg07661704) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:5.68E-06; Z-score:-8.84E-01 | ||
|
Methylation in Case |
6.11E-01 (Median) | Methylation in Control | 6.56E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC7A11 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
Body (cg04474257) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:1.11E-05; Z-score:-1.06E+00 | ||
|
Methylation in Case |
6.59E-01 (Median) | Methylation in Control | 7.20E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC7A11 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
Body (cg15867963) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:1.34E-05; Z-score:-1.94E+00 | ||
|
Methylation in Case |
6.27E-01 (Median) | Methylation in Control | 7.10E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC7A11 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
Body (cg06206831) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.42E-05; Z-score:-8.87E-01 | ||
|
Methylation in Case |
8.33E-01 (Median) | Methylation in Control | 8.57E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC7A11 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
Body (cg01309945) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.50E-05; Z-score:-8.60E-01 | ||
|
Methylation in Case |
7.77E-01 (Median) | Methylation in Control | 8.07E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC7A11 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
Body (cg06623625) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:1.43E-04; Z-score:-6.03E-01 | ||
|
Methylation in Case |
7.00E-01 (Median) | Methylation in Control | 7.64E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC7A11 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
Body (cg15441006) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.36E-03; Z-score:-2.62E-01 | ||
|
Methylation in Case |
6.89E-01 (Median) | Methylation in Control | 7.23E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC7A11 in hepatocellular carcinoma | [ 4 ] | |||
|
Location |
Body (cg24869834) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.04E-02; Z-score:-3.20E-01 | ||
|
Methylation in Case |
5.90E-01 (Median) | Methylation in Control | 6.11E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
HIV infection |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC7A11 in HIV infection | [ 5 ] | |||
|
Location |
TSS1500 (cg02102889) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.05E-02; Z-score:-4.46E-01 | ||
|
Methylation in Case |
8.22E-01 (Median) | Methylation in Control | 8.44E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC7A11 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg07661704) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.27E+00 | Statistic Test | p-value:6.38E-10; Z-score:2.08E+00 | ||
|
Methylation in Case |
7.24E-01 (Median) | Methylation in Control | 5.69E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC7A11 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg24869834) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:1.97E-06; Z-score:-1.50E+00 | ||
|
Methylation in Case |
7.83E-01 (Median) | Methylation in Control | 8.63E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC7A11 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg21877274) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.71E-05; Z-score:-1.89E+00 | ||
|
Methylation in Case |
8.56E-01 (Median) | Methylation in Control | 9.06E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC7A11 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg06623625) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:9.46E-03; Z-score:-8.07E-01 | ||
|
Methylation in Case |
9.26E-01 (Median) | Methylation in Control | 9.41E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC7A11 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg06206831) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.18E-02; Z-score:-6.84E-01 | ||
|
Methylation in Case |
9.20E-01 (Median) | Methylation in Control | 9.32E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC7A11 in panic disorder | [ 6 ] | |||
|
Location |
TSS1500 (cg02102889) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-8.38E-01 | Statistic Test | p-value:3.71E-04; Z-score:-5.16E-01 | ||
|
Methylation in Case |
-8.51E-01 (Median) | Methylation in Control | -7.14E-01 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC7A11 in panic disorder | [ 6 ] | |||
|
Location |
Body (cg24869834) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-4.41E-01 | Statistic Test | p-value:1.08E-06; Z-score:-9.06E-01 | ||
|
Methylation in Case |
-5.96E-01 (Median) | Methylation in Control | -2.63E-01 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC7A11 in panic disorder | [ 6 ] | |||
|
Location |
Body (cg02734904) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:7.18E-04; Z-score:-5.25E-01 | ||
|
Methylation in Case |
7.46E-01 (Median) | Methylation in Control | 1.01E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC7A11 in panic disorder | [ 6 ] | |||
|
Location |
Body (cg04487857) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:2.21E-02; Z-score:5.65E-01 | ||
|
Methylation in Case |
2.47E+00 (Median) | Methylation in Control | 2.33E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC7A11 in prostate cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg11200963) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.36E+00 | Statistic Test | p-value:2.95E-02; Z-score:2.45E+00 | ||
|
Methylation in Case |
7.33E-01 (Median) | Methylation in Control | 5.37E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC7A11 in prostate cancer | [ 7 ] | |||
|
Location |
3'UTR (cg19926635) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:1.62E-02; Z-score:1.95E+00 | ||
|
Methylation in Case |
7.42E-01 (Median) | Methylation in Control | 6.55E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Systemic lupus erythematosus |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC7A11 in systemic lupus erythematosus | [ 8 ] | |||
|
Location |
TSS1500 (cg00361146) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:1.38E-03; Z-score:-2.21E-01 | ||
|
Methylation in Case |
8.24E-02 (Median) | Methylation in Control | 9.21E-02 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC7A11 in systemic lupus erythematosus | [ 8 ] | |||
|
Location |
TSS1500 (cg02102889) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:2.70E-02; Z-score:3.66E-01 | ||
|
Methylation in Case |
7.43E-01 (Median) | Methylation in Control | 7.00E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC7A11 in systemic lupus erythematosus | [ 8 ] | |||
|
Location |
Body (cg07661704) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:2.08E-07; Z-score:5.52E-01 | ||
|
Methylation in Case |
6.34E-01 (Median) | Methylation in Control | 5.94E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC7A11 in systemic lupus erythematosus | [ 8 ] | |||
|
Location |
Body (cg01309945) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.29E-03; Z-score:-1.95E-01 | ||
|
Methylation in Case |
8.95E-01 (Median) | Methylation in Control | 9.00E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC7A11 in systemic lupus erythematosus | [ 8 ] | |||
|
Location |
Body (cg06623625) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:2.67E-02; Z-score:-1.57E-01 | ||
|
Methylation in Case |
9.18E-01 (Median) | Methylation in Control | 9.21E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC7A11 in systemic lupus erythematosus | [ 8 ] | |||
|
Location |
Body (cg21877274) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.72E-02; Z-score:-1.61E-01 | ||
|
Methylation in Case |
8.76E-01 (Median) | Methylation in Control | 8.87E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
8 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC7A11 in bladder cancer | [ 9 ] | |||
|
Location |
TSS200 (cg13028471) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.33E+00 | Statistic Test | p-value:5.55E-03; Z-score:-2.15E+00 | ||
|
Methylation in Case |
3.83E-02 (Median) | Methylation in Control | 8.90E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC7A11 in bladder cancer | [ 9 ] | |||
|
Location |
Body (cg15867963) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.65E+00 | Statistic Test | p-value:6.57E-07; Z-score:5.61E+00 | ||
|
Methylation in Case |
7.30E-01 (Median) | Methylation in Control | 4.42E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC7A11 in bladder cancer | [ 9 ] | |||
|
Location |
Body (cg06690548) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.57E+00 | Statistic Test | p-value:6.76E-07; Z-score:-5.59E+00 | ||
|
Methylation in Case |
3.93E-01 (Median) | Methylation in Control | 6.16E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC7A11 in bladder cancer | [ 9 ] | |||
|
Location |
Body (cg24869834) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.25E+00 | Statistic Test | p-value:2.06E-05; Z-score:-7.07E+00 | ||
|
Methylation in Case |
6.96E-01 (Median) | Methylation in Control | 8.73E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC7A11 in bladder cancer | [ 9 ] | |||
|
Location |
Body (cg02734904) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.16E-03; Z-score:-3.93E+00 | ||
|
Methylation in Case |
8.83E-01 (Median) | Methylation in Control | 9.16E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC7A11 in bladder cancer | [ 9 ] | |||
|
Location |
Body (cg04487857) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.92E+00 | Statistic Test | p-value:1.81E-03; Z-score:-3.67E+00 | ||
|
Methylation in Case |
1.97E-01 (Median) | Methylation in Control | 3.78E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC7A11 in bladder cancer | [ 9 ] | |||
|
Location |
Body (cg06206831) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:3.66E-03; Z-score:-2.16E+00 | ||
|
Methylation in Case |
8.83E-01 (Median) | Methylation in Control | 9.08E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC7A11 in bladder cancer | [ 9 ] | |||
|
Location |
Body (cg21877274) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:9.83E-03; Z-score:-1.95E+00 | ||
|
Methylation in Case |
8.38E-01 (Median) | Methylation in Control | 8.69E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC7A11 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
TSS200 (cg13028471) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.00E+00 | Statistic Test | p-value:1.44E-03; Z-score:-1.77E+00 | ||
|
Methylation in Case |
1.06E-01 (Median) | Methylation in Control | 2.12E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC7A11 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg21877274) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.74E-03; Z-score:-1.85E+00 | ||
|
Methylation in Case |
8.23E-01 (Median) | Methylation in Control | 8.70E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC7A11 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg24869834) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:5.63E-03; Z-score:-2.74E+00 | ||
|
Methylation in Case |
7.60E-01 (Median) | Methylation in Control | 8.32E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC7A11 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg06623625) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:2.46E-02; Z-score:9.92E-01 | ||
|
Methylation in Case |
8.74E-01 (Median) | Methylation in Control | 8.47E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC7A11 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg04474257) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:3.49E-02; Z-score:-2.06E+00 | ||
|
Methylation in Case |
7.89E-01 (Median) | Methylation in Control | 8.28E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
8 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC7A11 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
TSS200 (cg13028471) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:2.70E-06; Z-score:-7.00E-01 | ||
|
Methylation in Case |
5.14E-02 (Median) | Methylation in Control | 5.80E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC7A11 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg04487857) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.62E+00 | Statistic Test | p-value:3.83E-11; Z-score:2.00E+00 | ||
|
Methylation in Case |
3.43E-01 (Median) | Methylation in Control | 2.12E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC7A11 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg15441006) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:2.06E-08; Z-score:-1.46E+00 | ||
|
Methylation in Case |
7.89E-01 (Median) | Methylation in Control | 8.23E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC7A11 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg06623625) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:7.43E-07; Z-score:1.57E+00 | ||
|
Methylation in Case |
9.07E-01 (Median) | Methylation in Control | 8.59E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC7A11 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg15867963) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.31E+00 | Statistic Test | p-value:1.21E-06; Z-score:1.34E+00 | ||
|
Methylation in Case |
4.16E-01 (Median) | Methylation in Control | 3.17E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC7A11 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg24869834) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.15E-03; Z-score:-7.38E-01 | ||
|
Methylation in Case |
8.69E-01 (Median) | Methylation in Control | 8.92E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC7A11 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg07661704) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:1.28E-02; Z-score:5.33E-01 | ||
|
Methylation in Case |
8.13E-01 (Median) | Methylation in Control | 7.95E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC7A11 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg06206831) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.36E-02; Z-score:-5.09E-01 | ||
|
Methylation in Case |
9.46E-01 (Median) | Methylation in Control | 9.51E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Atypical teratoid rhabdoid tumor |
8 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC7A11 in atypical teratoid rhabdoid tumor | [ 12 ] | |||
|
Location |
Body (cg01309945) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.24E+00 | Statistic Test | p-value:6.08E-05; Z-score:-1.05E+00 | ||
|
Methylation in Case |
4.03E-01 (Median) | Methylation in Control | 5.01E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC7A11 in atypical teratoid rhabdoid tumor | [ 12 ] | |||
|
Location |
Body (cg02734904) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.49E+00 | Statistic Test | p-value:2.23E-04; Z-score:-1.06E+00 | ||
|
Methylation in Case |
2.18E-01 (Median) | Methylation in Control | 3.25E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC7A11 in atypical teratoid rhabdoid tumor | [ 12 ] | |||
|
Location |
Body (cg04474257) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.39E+00 | Statistic Test | p-value:7.96E-04; Z-score:-1.11E+00 | ||
|
Methylation in Case |
3.33E-01 (Median) | Methylation in Control | 4.61E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC7A11 in atypical teratoid rhabdoid tumor | [ 12 ] | |||
|
Location |
Body (cg04487857) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:8.25E-04; Z-score:-5.86E-01 | ||
|
Methylation in Case |
8.54E-01 (Median) | Methylation in Control | 8.79E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC7A11 in atypical teratoid rhabdoid tumor | [ 12 ] | |||
|
Location |
Body (cg06206831) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.31E+00 | Statistic Test | p-value:2.24E-03; Z-score:-5.75E-01 | ||
|
Methylation in Case |
2.75E-01 (Median) | Methylation in Control | 3.60E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC7A11 in atypical teratoid rhabdoid tumor | [ 12 ] | |||
|
Location |
Body (cg06623625) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:2.69E-03; Z-score:7.31E-01 | ||
|
Methylation in Case |
9.29E-01 (Median) | Methylation in Control | 8.35E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC7A11 in atypical teratoid rhabdoid tumor | [ 12 ] | |||
|
Location |
Body (cg06690548) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:2.81E-03; Z-score:-6.57E-01 | ||
|
Methylation in Case |
5.47E-01 (Median) | Methylation in Control | 6.27E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
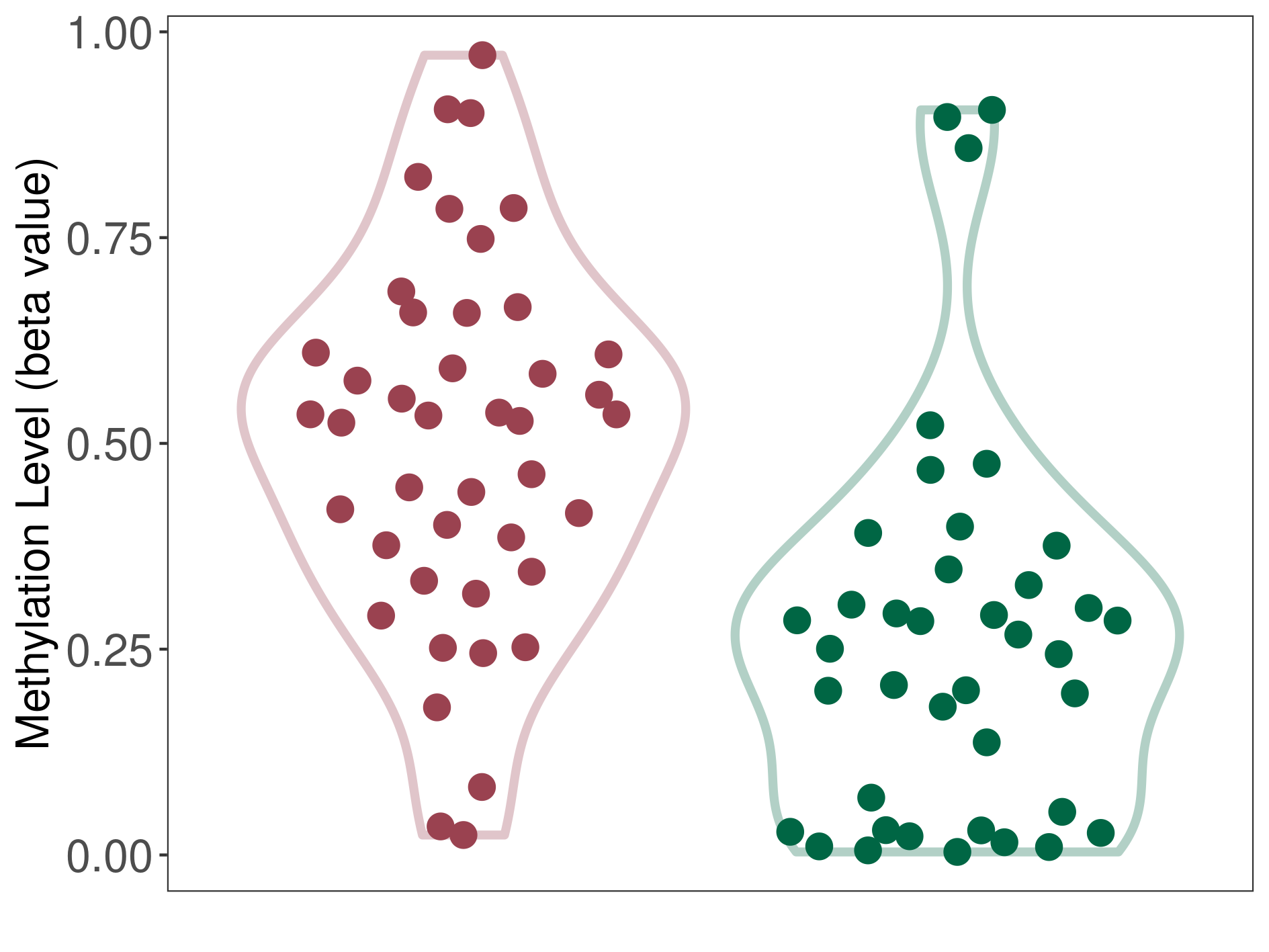
Epigenetic Phenomenon8 |
Methylation of SLC7A11 in atypical teratoid rhabdoid tumor | [ 12 ] | |||
|
Location |
Body (cg07661704) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:4.69E-03; Z-score:4.54E-01 | ||
|
Methylation in Case |
8.54E-01 (Median) | Methylation in Control | 8.20E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
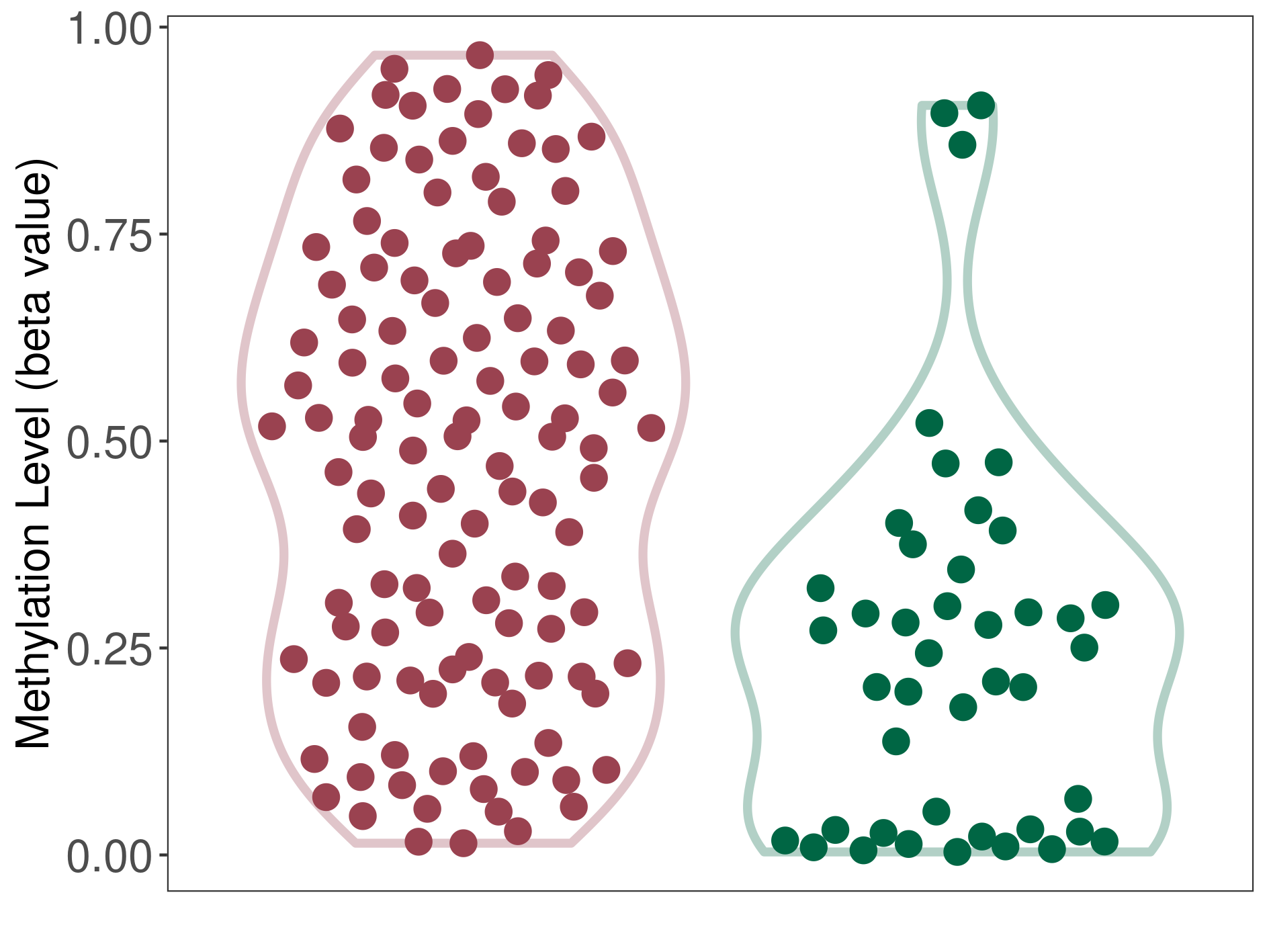
Atypical teratoid rhabdoid tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypermethylation of SLC7A11 in atypical teratoid rhabdoid tumour than that in healthy individual | ||||
Studied Phenotype |
Atypical teratoid rhabdoid tumour [ICD-11:2A00.1Y] | ||||
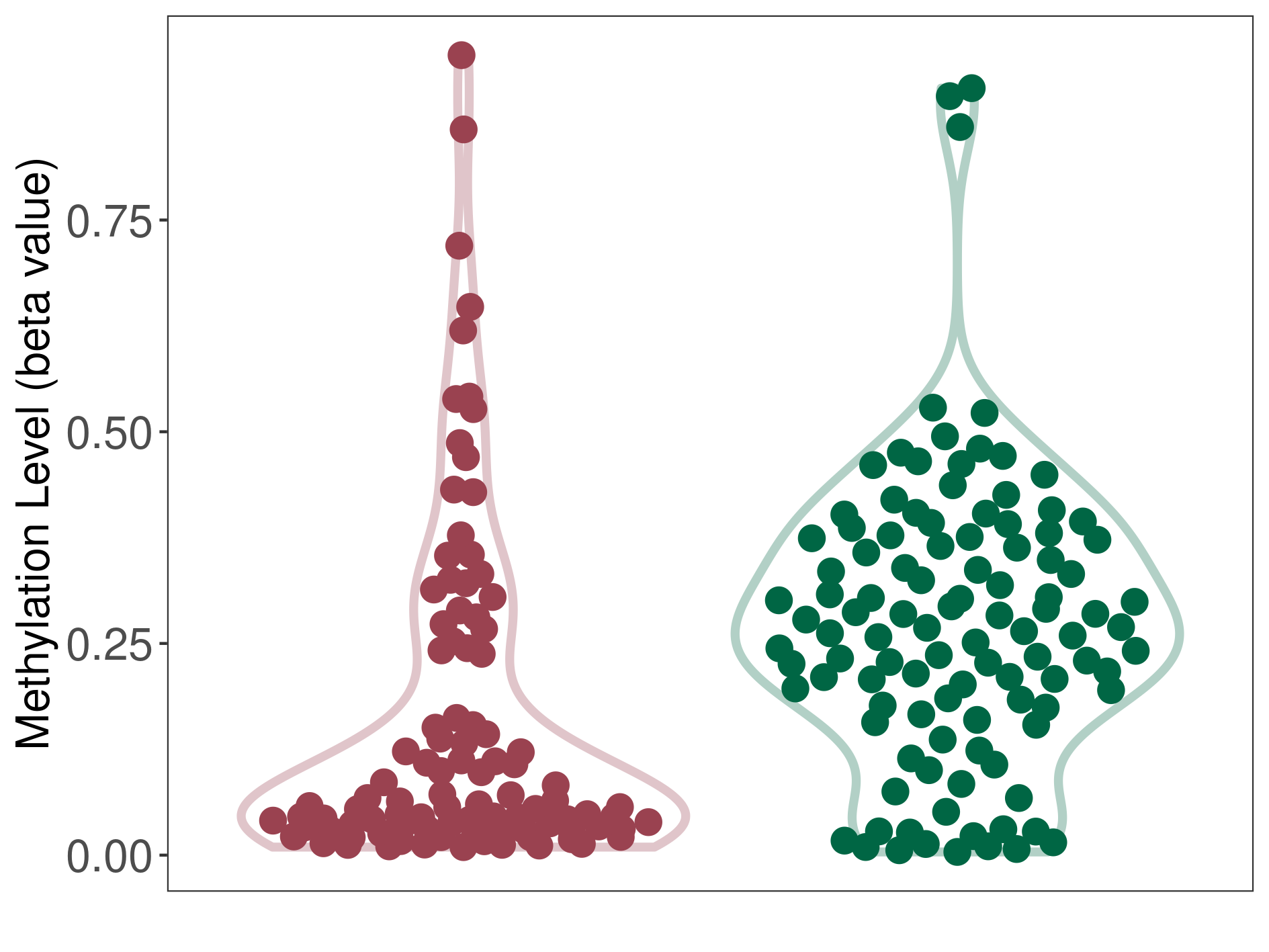
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:8.25E-07; Fold-change:0.261444373; Z-score:1.115094393 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
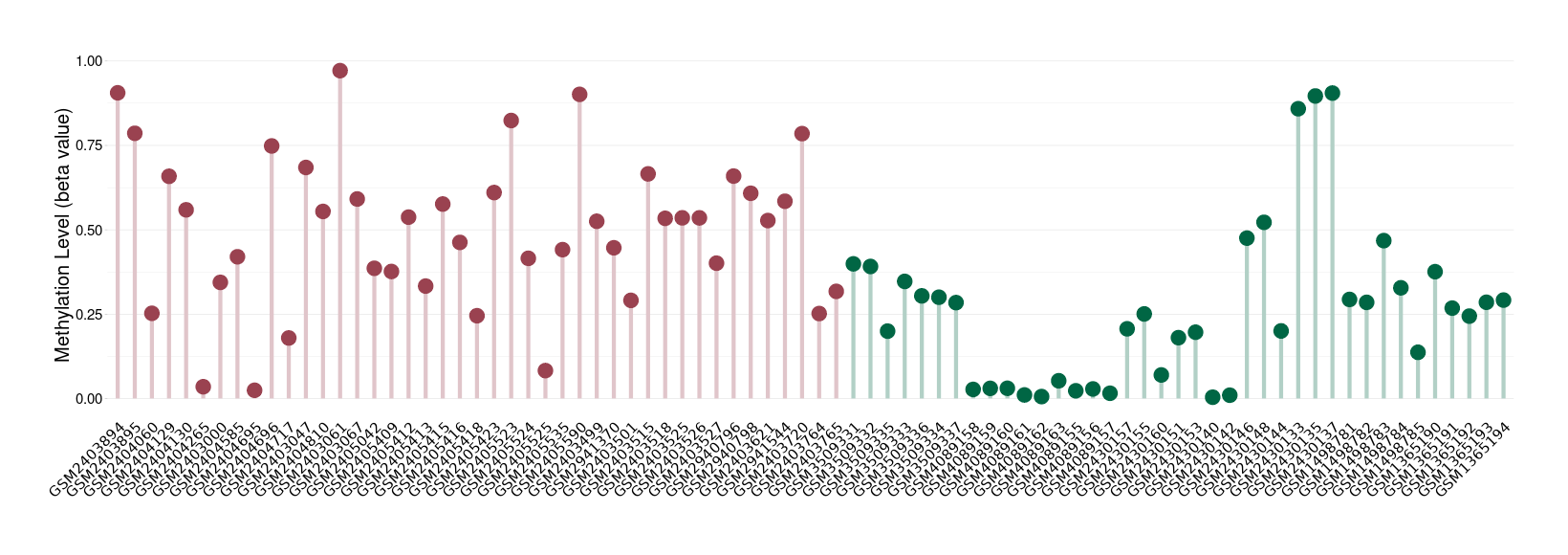
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Brain neuroblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypermethylation of SLC7A11 in brain neuroblastoma than that in healthy individual | ||||
Studied Phenotype |
Brain neuroblastoma [ICD-11:2A00.11] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.08E-05; Fold-change:0.283373055; Z-score:1.202963567 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Ovarian cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC7A11 in ovarian cancer than that in healthy individual | ||||
Studied Phenotype |
Ovarian cancer [ICD-11:2C73] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.013425961; Fold-change:-0.231306815; Z-score:-1.638472872 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
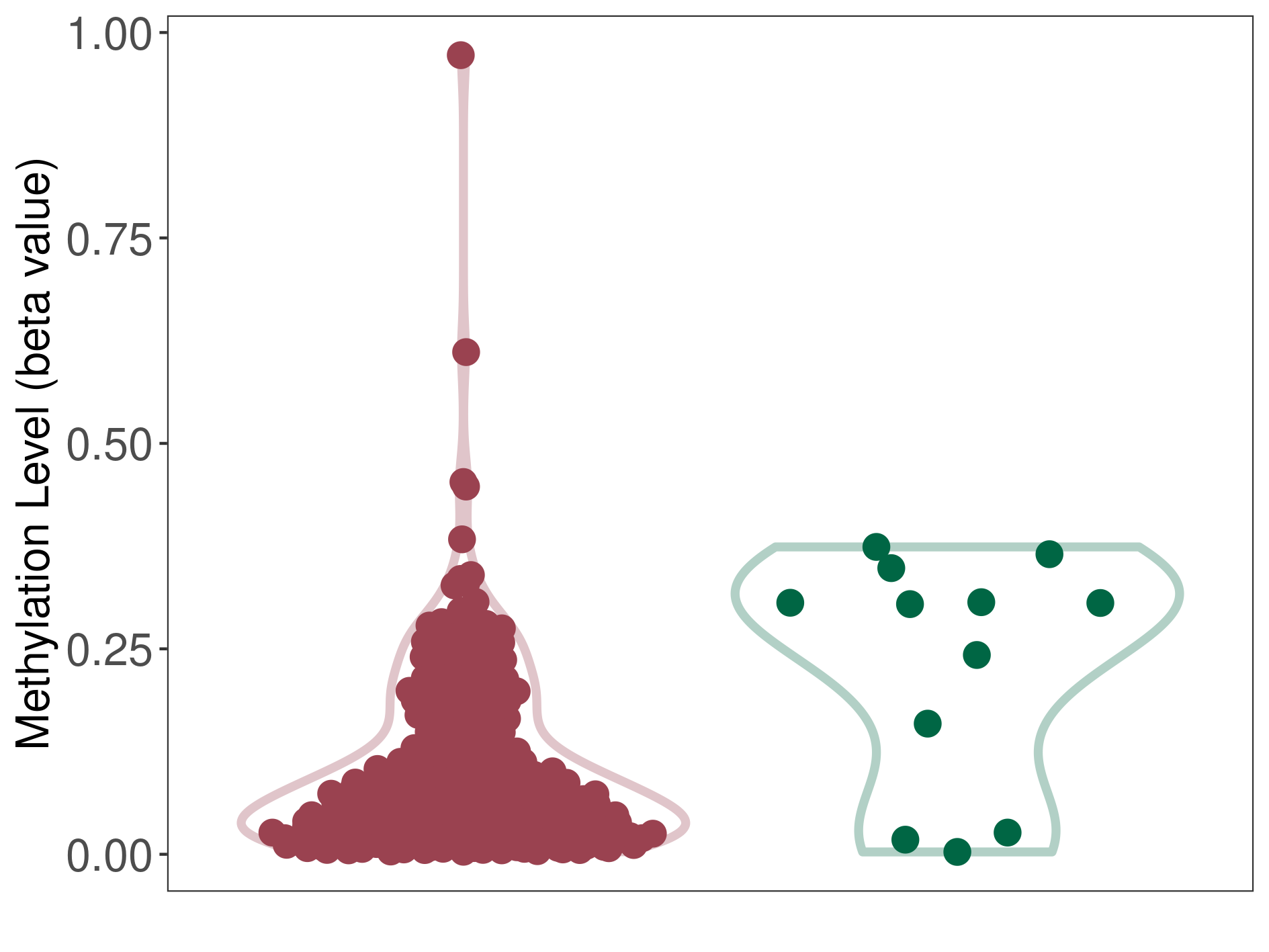
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
RELA YAP fusion ependymoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC7A11 in rela yap fusion ependymoma than that in healthy individual | ||||
Studied Phenotype |
RELA YAP fusion ependymoma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:2.98E-05; Fold-change:-0.206680629; Z-score:-1.196204455 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Cerebellar liponeurocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC7A11 in cerebellar liponeurocytoma than that in healthy individual | ||||
Studied Phenotype |
Cerebellar liponeurocytoma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.59E-09; Fold-change:0.742532468; Z-score:4.38326707 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||

|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Medulloblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC7A11 in medulloblastoma than that in healthy individual | ||||
Studied Phenotype |
Medulloblastoma [ICD-11:2A00.10] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.78E-79; Fold-change:0.659515713; Z-score:4.783181087 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||

|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Melanocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC7A11 in melanocytoma than that in healthy individual | ||||
Studied Phenotype |
Melanocytoma [ICD-11:2F36.2] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.36E-08; Fold-change:0.434857324; Z-score:2.742889307 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||

|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Multilayered rosettes embryonal tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
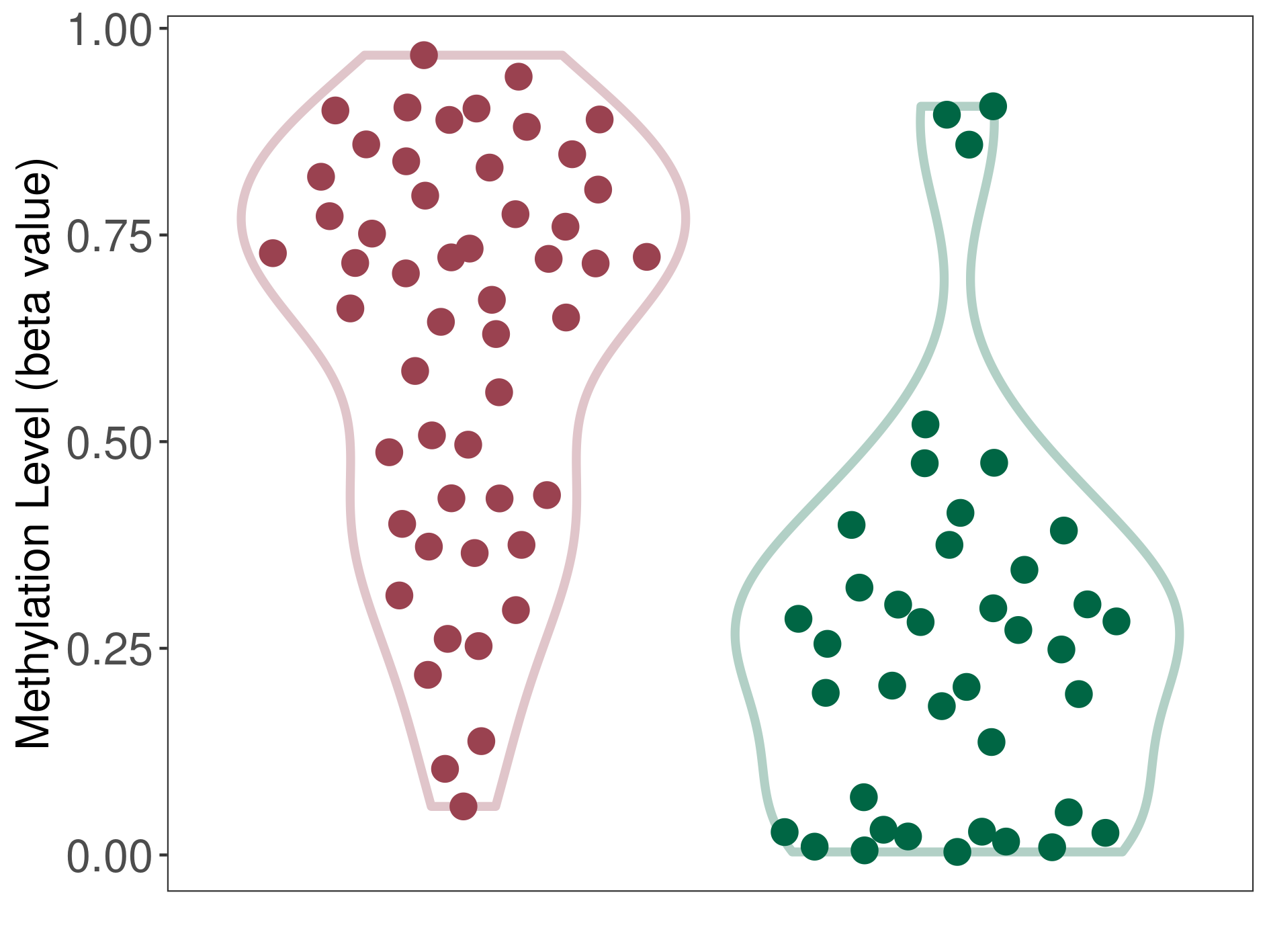
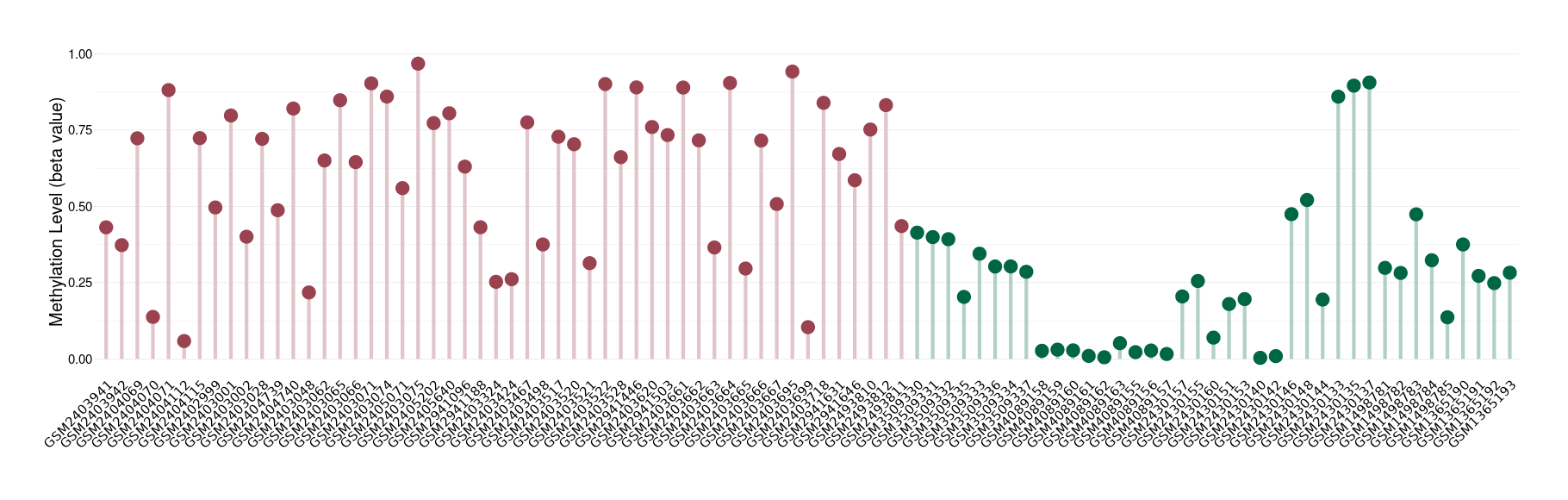
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC7A11 in multilayered rosettes embryonal tumour than that in healthy individual | ||||
Studied Phenotype |
Multilayered rosettes embryonal tumour [ICD-11:2A00.1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:5.66E-10; Fold-change:0.454182284; Z-score:1.916855181 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Oligodendroglioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC7A11 in oligodendroglioma than that in healthy individual | ||||
Studied Phenotype |
Oligodendroglioma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:4.88E-17; Fold-change:0.34605996; Z-score:1.714338 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||

|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Third ventricle chordoid glioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC7A11 in third ventricle chordoid glioma than that in healthy individual | ||||
Studied Phenotype |
Third ventricle chordoid glioma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.002176722; Fold-change:0.379485646; Z-score:2.569119328 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||

|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Lung cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC7A11 in lung cancer than that in adjacent tissue | ||||
Studied Phenotype |
Lung cancer [ICD-11:2C25] | ||||
The Methylation Level of Disease Section Compare with the Adjacent Tissue |
p-value:0.00234939; Fold-change:-0.230223429; Z-score:-1.916233374 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
DT methylation level in tissue other than the diseased tissue of patients
|
|||||
|
microRNA |
|||||
|
Bladder cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Higher expression of miR-27a in bladder cancer (compare with cisplatin-resistant counterpart cells) | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Dual-luciferase reporter assay | ||
|
Related Molecular Changes | Down regulation ofSLC7A11 | Experiment Method | Western Blot | ||
|
miRNA Stemloop ID |
miR-27a | miRNA Mature ID | miR-27a-3p | ||
|
miRNA Sequence |
UUCACAGUGGCUAAGUUCCGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
miR-27a overexpression may increase the sensitivity of bladder cancer cells to cisplatin by targeting SLC7A11. | ||||
|
Unclear Phenotype |
95 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
miR-1 directly targets SLC7A11 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1 | miRNA Mature ID | miR-1-5p | ||
|
miRNA Sequence |
ACAUACUUCUUUAUAUGCCCAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon2 |
miR-106a directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-106a | miRNA Mature ID | miR-106a-5p | ||
|
miRNA Sequence |
AAAAGUGCUUACAGUGCAGGUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon3 |
miR-106b directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-106b | miRNA Mature ID | miR-106b-5p | ||
|
miRNA Sequence |
UAAAGUGCUGACAGUGCAGAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon4 |
miR-1247 directly targets SLC7A11 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-1247 | miRNA Mature ID | miR-1247-3p | ||
|
miRNA Sequence |
CCCCGGGAACGUCGAGACUGGAGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon5 |
miR-1277 directly targets SLC7A11 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-1277 | miRNA Mature ID | miR-1277-5p | ||
|
miRNA Sequence |
AAAUAUAUAUAUAUAUGUACGUAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon6 |
miR-1279 directly targets SLC7A11 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1279 | miRNA Mature ID | miR-1279 | ||
|
miRNA Sequence |
UCAUAUUGCUUCUUUCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon7 |
miR-128 directly targets SLC7A11 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-128 | miRNA Mature ID | miR-128-3p | ||
|
miRNA Sequence |
UCACAGUGAACCGGUCUCUUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human cervical cancer cell line (Hela) | ||||
|
Epigenetic Phenomenon8 |
miR-1281 directly targets SLC7A11 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-1281 | miRNA Mature ID | miR-1281 | ||
|
miRNA Sequence |
UCGCCUCCUCCUCUCCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon9 |
miR-142 directly targets SLC7A11 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-142 | miRNA Mature ID | miR-142-3p | ||
|
miRNA Sequence |
UGUAGUGUUUCCUACUUUAUGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon10 |
miR-148b directly targets SLC7A11 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-148b | miRNA Mature ID | miR-148b-3p | ||
|
miRNA Sequence |
UCAGUGCAUCACAGAACUUUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human cervical cancer cell line (Hela) | ||||
|
Epigenetic Phenomenon11 |
miR-150 directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-150 | miRNA Mature ID | miR-150-5p | ||
|
miRNA Sequence |
UCUCCCAACCCUUGUACCAGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon12 |
miR-155 directly targets SLC7A11 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Proteomics | ||
|
miRNA Stemloop ID |
miR-155 | miRNA Mature ID | miR-155-5p | ||
|
miRNA Sequence |
UUAAUGCUAAUCGUGAUAGGGGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human cervical cancer cell line (Hela) | ||||
|
Epigenetic Phenomenon13 |
miR-17 directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-17 | miRNA Mature ID | miR-17-5p | ||
|
miRNA Sequence |
CAAAGUGCUUACAGUGCAGGUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon14 |
miR-181a directly targets SLC7A11 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-181a | miRNA Mature ID | miR-181a-5p | ||
|
miRNA Sequence |
AACAUUCAACGCUGUCGGUGAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human cervical cancer cell line (Hela) | ||||
|
Epigenetic Phenomenon15 |
miR-186 directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-186 | miRNA Mature ID | miR-186-3p | ||
|
miRNA Sequence |
GCCCAAAGGUGAAUUUUUUGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon16 |
miR-190a directly targets SLC7A11 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-190a | miRNA Mature ID | miR-190a-3p | ||
|
miRNA Sequence |
CUAUAUAUCAAACAUAUUCCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon17 |
miR-192 directly targets SLC7A11 | [ 22 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-192 | miRNA Mature ID | miR-192-5p | ||
|
miRNA Sequence |
CUGACCUAUGAAUUGACAGCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon18 |
miR-1976 directly targets SLC7A11 | [ 23 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1976 | miRNA Mature ID | miR-1976 | ||
|
miRNA Sequence |
CCUCCUGCCCUCCUUGCUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon19 |
miR-19a directly targets SLC7A11 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-19a | miRNA Mature ID | miR-19a-3p | ||
|
miRNA Sequence |
UGUGCAAAUCUAUGCAAAACUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon20 |
miR-19b directly targets SLC7A11 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-19b | miRNA Mature ID | miR-19b-3p | ||
|
miRNA Sequence |
UGUGCAAAUCCAUGCAAAACUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon21 |
miR-20a directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-20a | miRNA Mature ID | miR-20a-5p | ||
|
miRNA Sequence |
UAAAGUGCUUAUAGUGCAGGUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon22 |
miR-20b directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-20b | miRNA Mature ID | miR-20b-5p | ||
|
miRNA Sequence |
CAAAGUGCUCAUAGUGCAGGUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon23 |
miR-215 directly targets SLC7A11 | [ 22 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-215 | miRNA Mature ID | miR-215-5p | ||
|
miRNA Sequence |
AUGACCUAUGAAUUGACAGAC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon24 |
miR-218 directly targets SLC7A11 | [ 24 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-218 | miRNA Mature ID | miR-218-5p | ||
|
miRNA Sequence |
UUGUGCUUGAUCUAACCAUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon25 |
miR-223 directly targets SLC7A11 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-223 | miRNA Mature ID | miR-223-5p | ||
|
miRNA Sequence |
CGUGUAUUUGACAAGCUGAGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon26 |
miR-25 directly targets SLC7A11 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-25 | miRNA Mature ID | miR-25-3p | ||
|
miRNA Sequence |
CAUUGCACUUGUCUCGGUCUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon27 |
miR-26b directly targets SLC7A11 | [ 25 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Sequencing | ||
|
miRNA Stemloop ID |
miR-26b | miRNA Mature ID | miR-26b-5p | ||
|
miRNA Sequence |
UUCAAGUAAUUCAGGAUAGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon28 |
miR-27a directly targets SLC7A11 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Immunohistochemistry//Luciferase reporter assay//qRT-PCR | ||
|
miRNA Stemloop ID |
miR-27a | miRNA Mature ID | miR-27a-3p | ||
|
miRNA Sequence |
UUCACAGUGGCUAAGUUCCGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon29 |
miR-302a directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-302a | miRNA Mature ID | miR-302a-3p | ||
|
miRNA Sequence |
UAAGUGCUUCCAUGUUUUGGUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon30 |
miR-302b directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-302b | miRNA Mature ID | miR-302b-3p | ||
|
miRNA Sequence |
UAAGUGCUUCCAUGUUUUAGUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon31 |
miR-302c directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-302c | miRNA Mature ID | miR-302c-3p | ||
|
miRNA Sequence |
UAAGUGCUUCCAUGUUUCAGUGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon32 |
miR-302d directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-302d | miRNA Mature ID | miR-302d-3p | ||
|
miRNA Sequence |
UAAGUGCUUCCAUGUUUGAGUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon33 |
miR-302e directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-302e | miRNA Mature ID | miR-302e | ||
|
miRNA Sequence |
UAAGUGCUUCCAUGCUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon34 |
miR-30a directly targets SLC7A11 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | pSILAC//Proteomics;Other | ||
|
miRNA Stemloop ID |
miR-30a | miRNA Mature ID | miR-30a-5p | ||
|
miRNA Sequence |
UGUAAACAUCCUCGACUGGAAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human cervical cancer cell line (Hela) | ||||
|
Epigenetic Phenomenon35 |
miR-3163 directly targets SLC7A11 | [ 25 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3163 | miRNA Mature ID | miR-3163 | ||
|
miRNA Sequence |
UAUAAAAUGAGGGCAGUAAGAC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon36 |
miR-32 directly targets SLC7A11 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-32 | miRNA Mature ID | miR-32-5p | ||
|
miRNA Sequence |
UAUUGCACAUUACUAAGUUGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon37 |
miR-329 directly targets SLC7A11 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-329 | miRNA Mature ID | miR-329-3p | ||
|
miRNA Sequence |
AACACACCUGGUUAACCUCUUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon38 |
miR-340 directly targets SLC7A11 | [ 25 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Sequencing | ||
|
miRNA Stemloop ID |
miR-340 | miRNA Mature ID | miR-340-5p | ||
|
miRNA Sequence |
UUAUAAAGCAAUGAGACUGAUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon39 |
miR-362 directly targets SLC7A11 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-362 | miRNA Mature ID | miR-362-3p | ||
|
miRNA Sequence |
AACACACCUAUUCAAGGAUUCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon40 |
miR-363 directly targets SLC7A11 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-363 | miRNA Mature ID | miR-363-3p | ||
|
miRNA Sequence |
AAUUGCACGGUAUCCAUCUGUA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon41 |
miR-367 directly targets SLC7A11 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-367 | miRNA Mature ID | miR-367-3p | ||
|
miRNA Sequence |
AAUUGCACUUUAGCAAUGGUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon42 |
miR-372 directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-372 | miRNA Mature ID | miR-372-3p | ||
|
miRNA Sequence |
AAAGUGCUGCGACAUUUGAGCGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon43 |
miR-373 directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-373 | miRNA Mature ID | miR-373-3p | ||
|
miRNA Sequence |
GAAGUGCUUCGAUUUUGGGGUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon44 |
miR-3913 directly targets SLC7A11 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3913 | miRNA Mature ID | miR-3913-3p | ||
|
miRNA Sequence |
AGACAUCAAGAUCAGUCCCAAA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon45 |
miR-3941 directly targets SLC7A11 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3941 | miRNA Mature ID | miR-3941 | ||
|
miRNA Sequence |
UUACACACAACUGAGGAUCAUA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon46 |
miR-410 directly targets SLC7A11 | [ 25 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-410 | miRNA Mature ID | miR-410-3p | ||
|
miRNA Sequence |
AAUAUAACACAGAUGGCCUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon47 |
miR-4279 directly targets SLC7A11 | [ 23 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4279 | miRNA Mature ID | miR-4279 | ||
|
miRNA Sequence |
CUCUCCUCCCGGCUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon48 |
miR-4282 directly targets SLC7A11 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4282 | miRNA Mature ID | miR-4282 | ||
|
miRNA Sequence |
UAAAAUUUGCAUCCAGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon49 |
miR-4640 directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4640 | miRNA Mature ID | miR-4640-3p | ||
|
miRNA Sequence |
CACCCCCUGUUUCCUGGCCCAC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon50 |
miR-4722 directly targets SLC7A11 | [ 23 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4722 | miRNA Mature ID | miR-4722-3p | ||
|
miRNA Sequence |
ACCUGCCAGCACCUCCCUGCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon51 |
miR-4731 directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4731 | miRNA Mature ID | miR-4731-5p | ||
|
miRNA Sequence |
UGCUGGGGGCCACAUGAGUGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon52 |
miR-4789 directly targets SLC7A11 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4789 | miRNA Mature ID | miR-4789-3p | ||
|
miRNA Sequence |
CACACAUAGCAGGUGUAUAUA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon53 |
miR-4789 directly targets SLC7A11 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4789 | miRNA Mature ID | miR-4789-5p | ||
|
miRNA Sequence |
GUAUACACCUGAUAUGUGUAUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon54 |
miR-489 directly targets SLC7A11 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-489 | miRNA Mature ID | miR-489-3p | ||
|
miRNA Sequence |
GUGACAUCACAUAUACGGCAGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon55 |
miR-500a directly targets SLC7A11 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-500a | miRNA Mature ID | miR-500a-3p | ||
|
miRNA Sequence |
AUGCACCUGGGCAAGGAUUCUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon56 |
miR-5011 directly targets SLC7A11 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5011 | miRNA Mature ID | miR-5011-5p | ||
|
miRNA Sequence |
UAUAUAUACAGCCAUGCACUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon57 |
miR-505 directly targets SLC7A11 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-505 | miRNA Mature ID | miR-505-5p | ||
|
miRNA Sequence |
GGGAGCCAGGAAGUAUUGAUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon58 |
miR-5089 directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5089 | miRNA Mature ID | miR-5089-5p | ||
|
miRNA Sequence |
GUGGGAUUUCUGAGUAGCAUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon59 |
miR-512 directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-512 | miRNA Mature ID | miR-512-3p | ||
|
miRNA Sequence |
AAGUGCUGUCAUAGCUGAGGUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon60 |
miR-5193 directly targets SLC7A11 | [ 23 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-5193 | miRNA Mature ID | miR-5193 | ||
|
miRNA Sequence |
UCCUCCUCUACCUCAUCCCAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon61 |
miR-519d directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-519d | miRNA Mature ID | miR-519d-3p | ||
|
miRNA Sequence |
CAAAGUGCCUCCCUUUAGAGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon62 |
miR-520a directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-520a | miRNA Mature ID | miR-520a-3p | ||
|
miRNA Sequence |
AAAGUGCUUCCCUUUGGACUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon63 |
miR-520c directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-520c | miRNA Mature ID | miR-520c-3p | ||
|
miRNA Sequence |
AAAGUGCUUCCUUUUAGAGGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon64 |
miR-520d directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-520d | miRNA Mature ID | miR-520d-3p | ||
|
miRNA Sequence |
AAAGUGCUUCUCUUUGGUGGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon65 |
miR-520g directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-520g | miRNA Mature ID | miR-520g-3p | ||
|
miRNA Sequence |
ACAAAGUGCUUCCCUUUAGAGUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon66 |
miR-520h directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-520h | miRNA Mature ID | miR-520h | ||
|
miRNA Sequence |
ACAAAGUGCUUCCCUUUAGAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon67 |
miR-526b directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-526b | miRNA Mature ID | miR-526b-3p | ||
|
miRNA Sequence |
GAAAGUGCUUCCUUUUAGAGGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon68 |
miR-548az directly targets SLC7A11 | [ 25 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-548az | miRNA Mature ID | miR-548az-5p | ||
|
miRNA Sequence |
CAAAAGUGAUUGUGGUUUUUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon69 |
miR-548e directly targets SLC7A11 | [ 25 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-548e | miRNA Mature ID | miR-548e-5p | ||
|
miRNA Sequence |
CAAAAGCAAUCGCGGUUUUUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon70 |
miR-548t directly targets SLC7A11 | [ 25 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-548t | miRNA Mature ID | miR-548t-5p | ||
|
miRNA Sequence |
CAAAAGUGAUCGUGGUUUUUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon71 |
miR-5571 directly targets SLC7A11 | [ 23 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-5571 | miRNA Mature ID | miR-5571-5p | ||
|
miRNA Sequence |
CAAUUCUCAAAGGAGCCUCCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon72 |
miR-5589 directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5589 | miRNA Mature ID | miR-5589-5p | ||
|
miRNA Sequence |
GGCUGGGUGCUCUUGUGCAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon73 |
miR-5589 directly targets SLC7A11 | [ 25 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-5589 | miRNA Mature ID | miR-5589-3p | ||
|
miRNA Sequence |
UGCACAUGGCAACCUAGCUCCCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon74 |
miR-5683 directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5683 | miRNA Mature ID | miR-5683 | ||
|
miRNA Sequence |
UACAGAUGCAGAUUCUCUGACUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon75 |
miR-574 directly targets SLC7A11 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-574 | miRNA Mature ID | miR-574-5p | ||
|
miRNA Sequence |
UGAGUGUGUGUGUGUGAGUGUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon76 |
miR-587 directly targets SLC7A11 | [ 25 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-587 | miRNA Mature ID | miR-587 | ||
|
miRNA Sequence |
UUUCCAUAGGUGAUGAGUCAC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon77 |
miR-595 directly targets SLC7A11 | [ 25 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-595 | miRNA Mature ID | miR-595 | ||
|
miRNA Sequence |
GAAGUGUGCCGUGGUGUGUCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon78 |
miR-603 directly targets SLC7A11 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-603 | miRNA Mature ID | miR-603 | ||
|
miRNA Sequence |
CACACACUGCAAUUACUUUUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon79 |
miR-619 directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-619 | miRNA Mature ID | miR-619-5p | ||
|
miRNA Sequence |
GCUGGGAUUACAGGCAUGAGCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon80 |
miR-6504 directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6504 | miRNA Mature ID | miR-6504-3p | ||
|
miRNA Sequence |
CAUUACAGCACAGCCAUUCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon81 |
miR-6506 directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6506 | miRNA Mature ID | miR-6506-5p | ||
|
miRNA Sequence |
ACUGGGAUGUCACUGAAUAUGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon82 |
miR-6727 directly targets SLC7A11 | [ 23 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6727 | miRNA Mature ID | miR-6727-3p | ||
|
miRNA Sequence |
UCCUGCCACCUCCUCCGCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon83 |
miR-6747 directly targets SLC7A11 | [ 23 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6747 | miRNA Mature ID | miR-6747-3p | ||
|
miRNA Sequence |
UCCUGCCUUCCUCUGCACCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon84 |
miR-6778 directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6778 | miRNA Mature ID | miR-6778-3p | ||
|
miRNA Sequence |
UGCCUCCCUGACAUUCCACAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon85 |
miR-6791 directly targets SLC7A11 | [ 23 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6791 | miRNA Mature ID | miR-6791-3p | ||
|
miRNA Sequence |
UGCCUCCUUGGUCUCCGGCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon86 |
miR-6826 directly targets SLC7A11 | [ 23 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6826 | miRNA Mature ID | miR-6826-5p | ||
|
miRNA Sequence |
UCAAUAGGAAAGAGGUGGGACCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon87 |
miR-6829 directly targets SLC7A11 | [ 23 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6829 | miRNA Mature ID | miR-6829-3p | ||
|
miRNA Sequence |
UGCCUCCUCCGUGGCCUCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon88 |
miR-6835 directly targets SLC7A11 | [ 25 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6835 | miRNA Mature ID | miR-6835-3p | ||
|
miRNA Sequence |
AAAAGCACUUUUCUGUCUCCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon89 |
miR-6867 directly targets SLC7A11 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6867 | miRNA Mature ID | miR-6867-5p | ||
|
miRNA Sequence |
UGUGUGUGUAGAGGAAGAAGGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon90 |
miR-6874 directly targets SLC7A11 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6874 | miRNA Mature ID | miR-6874-5p | ||
|
miRNA Sequence |
AUGGAGCUGGAACCAGAUCAGGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon91 |
miR-767 directly targets SLC7A11 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-767 | miRNA Mature ID | miR-767-5p | ||
|
miRNA Sequence |
UGCACCAUGGUUGUCUGAGCAUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon92 |
miR-8485 directly targets SLC7A11 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-8485 | miRNA Mature ID | miR-8485 | ||
|
miRNA Sequence |
CACACACACACACACACGUAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon93 |
miR-92a directly targets SLC7A11 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-92a | miRNA Mature ID | miR-92a-3p | ||
|
miRNA Sequence |
UAUUGCACUUGUCCCGGCCUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon94 |
miR-92b directly targets SLC7A11 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-92b | miRNA Mature ID | miR-92b-3p | ||
|
miRNA Sequence |
UAUUGCACUCGUCCCGGCCUCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon95 |
miR-93 directly targets SLC7A11 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-93 | miRNA Mature ID | miR-93-5p | ||
|
miRNA Sequence |
CAAAGUGCUGUUCGUGCAGGUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
