Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0457 Transporter Info | ||||
| Gene Name | SLC6A5 | ||||
| Transporter Name | Sodium- and chloride-dependent glycine transporter 2 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Colon cancer |
20 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg13425422) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.29E+00 | Statistic Test | p-value:2.88E-07; Z-score:-5.61E+00 | ||
|
Methylation in Case |
6.56E-01 (Median) | Methylation in Control | 8.45E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg23677272) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:7.05E-06; Z-score:-3.18E+00 | ||
|
Methylation in Case |
5.28E-01 (Median) | Methylation in Control | 6.24E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg06246950) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.34E+00 | Statistic Test | p-value:1.72E-05; Z-score:-2.44E+00 | ||
|
Methylation in Case |
3.74E-01 (Median) | Methylation in Control | 5.02E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg13613146) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.20E+00 | Statistic Test | p-value:5.94E-05; Z-score:-1.57E+00 | ||
|
Methylation in Case |
5.35E-01 (Median) | Methylation in Control | 6.39E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg02828514) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.17E+00 | Statistic Test | p-value:9.48E-04; Z-score:-1.62E+00 | ||
|
Methylation in Case |
4.71E-01 (Median) | Methylation in Control | 5.53E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg08074851) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.09E+00 | Statistic Test | p-value:1.46E-10; Z-score:3.88E+00 | ||
|
Methylation in Case |
5.34E-01 (Median) | Methylation in Control | 2.55E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg14494721) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.49E+00 | Statistic Test | p-value:6.41E-05; Z-score:-1.41E+00 | ||
|
Methylation in Case |
2.27E-01 (Median) | Methylation in Control | 3.39E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg15173134) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.67E+00 | Statistic Test | p-value:4.23E-04; Z-score:2.86E+00 | ||
|
Methylation in Case |
6.19E-01 (Median) | Methylation in Control | 3.70E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg21487099) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:4.73E-04; Z-score:-1.78E+00 | ||
|
Methylation in Case |
5.14E-01 (Median) | Methylation in Control | 5.69E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg01323777) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:1.47E-03; Z-score:6.84E-01 | ||
|
Methylation in Case |
8.89E-01 (Median) | Methylation in Control | 8.38E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
1stExon (cg26013553) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.38E+00 | Statistic Test | p-value:4.40E-07; Z-score:1.24E+00 | ||
|
Methylation in Case |
5.76E-01 (Median) | Methylation in Control | 4.16E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg13678205) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.40E+00 | Statistic Test | p-value:7.58E-08; Z-score:-3.44E+00 | ||
|
Methylation in Case |
5.24E-01 (Median) | Methylation in Control | 7.33E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg10737118) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.49E+00 | Statistic Test | p-value:2.97E-07; Z-score:3.51E+00 | ||
|
Methylation in Case |
4.27E-01 (Median) | Methylation in Control | 2.86E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg21961694) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.51E+00 | Statistic Test | p-value:8.35E-07; Z-score:-3.16E+00 | ||
|
Methylation in Case |
4.14E-01 (Median) | Methylation in Control | 6.27E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg01367751) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:2.02E-06; Z-score:-2.44E+00 | ||
|
Methylation in Case |
6.96E-01 (Median) | Methylation in Control | 8.10E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg00161124) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.47E+00 | Statistic Test | p-value:1.49E-05; Z-score:-2.64E+00 | ||
|
Methylation in Case |
3.12E-01 (Median) | Methylation in Control | 4.57E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg00445475) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:1.01E-04; Z-score:2.16E+00 | ||
|
Methylation in Case |
4.90E-01 (Median) | Methylation in Control | 4.18E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg18856043) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:1.63E-03; Z-score:-1.38E+00 | ||
|
Methylation in Case |
3.95E-01 (Median) | Methylation in Control | 4.48E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
3'UTR (cg21477176) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.31E+00 | Statistic Test | p-value:1.55E-06; Z-score:2.22E+00 | ||
|
Methylation in Case |
7.17E-01 (Median) | Methylation in Control | 5.48E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC6A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
3'UTR (cg22035738) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:2.43E-03; Z-score:-1.92E+00 | ||
|
Methylation in Case |
6.15E-01 (Median) | Methylation in Control | 6.88E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
36 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
5'UTR (cg02608452) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.64E+00 | Statistic Test | p-value:3.74E-18; Z-score:-3.52E+00 | ||
|
Methylation in Case |
3.77E-01 (Median) | Methylation in Control | 6.19E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg02313331) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.65E+00 | Statistic Test | p-value:6.88E-21; Z-score:2.73E+00 | ||
|
Methylation in Case |
6.03E-01 (Median) | Methylation in Control | 3.66E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg04433322) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.47E+00 | Statistic Test | p-value:1.87E-16; Z-score:-2.96E+00 | ||
|
Methylation in Case |
4.50E-01 (Median) | Methylation in Control | 6.62E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg08975046) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.73E+00 | Statistic Test | p-value:7.07E-15; Z-score:-3.44E+00 | ||
|
Methylation in Case |
4.12E-01 (Median) | Methylation in Control | 7.15E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg03573068) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.25E+00 | Statistic Test | p-value:1.35E-04; Z-score:1.42E+00 | ||
|
Methylation in Case |
2.46E-01 (Median) | Methylation in Control | 1.97E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg11288764) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:1.36E-03; Z-score:5.94E-01 | ||
|
Methylation in Case |
1.34E-01 (Median) | Methylation in Control | 1.21E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg09696301) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:4.12E-03; Z-score:-1.64E-01 | ||
|
Methylation in Case |
9.88E-02 (Median) | Methylation in Control | 1.02E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg11784785) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:6.00E-03; Z-score:2.48E-01 | ||
|
Methylation in Case |
1.10E-01 (Median) | Methylation in Control | 1.00E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg27036111) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:2.41E-02; Z-score:1.17E+00 | ||
|
Methylation in Case |
4.27E-01 (Median) | Methylation in Control | 3.75E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg24900654) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.30E+00 | Statistic Test | p-value:1.09E-11; Z-score:-2.78E+00 | ||
|
Methylation in Case |
4.36E-01 (Median) | Methylation in Control | 5.68E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg07645736) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.27E+00 | Statistic Test | p-value:5.19E-10; Z-score:3.51E+00 | ||
|
Methylation in Case |
1.70E-01 (Median) | Methylation in Control | 7.52E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg20551181) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.34E+00 | Statistic Test | p-value:6.35E-05; Z-score:8.33E-01 | ||
|
Methylation in Case |
1.73E-01 (Median) | Methylation in Control | 1.29E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg16635352) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:7.94E-05; Z-score:1.21E+00 | ||
|
Methylation in Case |
3.40E-01 (Median) | Methylation in Control | 2.83E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg06804921) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:1.86E-03; Z-score:3.78E-02 | ||
|
Methylation in Case |
6.48E-02 (Median) | Methylation in Control | 6.41E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg08530317) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:5.12E-03; Z-score:3.33E-01 | ||
|
Methylation in Case |
1.05E-01 (Median) | Methylation in Control | 9.45E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
1stExon (cg07067659) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.16E+01 | Statistic Test | p-value:1.43E-14; Z-score:3.84E+00 | ||
|
Methylation in Case |
3.13E-01 (Median) | Methylation in Control | 2.70E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg09433131) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.16E+00 | Statistic Test | p-value:1.30E-23; Z-score:-8.32E+00 | ||
|
Methylation in Case |
4.25E-01 (Median) | Methylation in Control | 9.16E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg02913918) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.79E+00 | Statistic Test | p-value:2.67E-21; Z-score:-8.81E+00 | ||
|
Methylation in Case |
4.58E-01 (Median) | Methylation in Control | 8.18E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg19264163) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.77E+00 | Statistic Test | p-value:1.30E-20; Z-score:-3.87E+00 | ||
|
Methylation in Case |
4.30E-01 (Median) | Methylation in Control | 7.62E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg17260954) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.90E+00 | Statistic Test | p-value:1.66E-19; Z-score:-1.03E+01 | ||
|
Methylation in Case |
4.39E-01 (Median) | Methylation in Control | 8.35E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg26269038) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.37E+00 | Statistic Test | p-value:3.21E-16; Z-score:-8.26E+00 | ||
|
Methylation in Case |
5.92E-01 (Median) | Methylation in Control | 8.12E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg07262519) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.43E+00 | Statistic Test | p-value:7.69E-16; Z-score:-3.99E+00 | ||
|
Methylation in Case |
5.03E-01 (Median) | Methylation in Control | 7.17E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg19930802) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.33E+00 | Statistic Test | p-value:5.06E-14; Z-score:-1.06E+01 | ||
|
Methylation in Case |
6.49E-01 (Median) | Methylation in Control | 8.63E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg08164556) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.32E+00 | Statistic Test | p-value:6.64E-11; Z-score:5.46E+00 | ||
|
Methylation in Case |
3.18E-01 (Median) | Methylation in Control | 1.38E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg20714059) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:5.29E-10; Z-score:-1.74E+00 | ||
|
Methylation in Case |
8.88E-01 (Median) | Methylation in Control | 9.25E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg09136245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.27E+00 | Statistic Test | p-value:6.45E-09; Z-score:-1.04E+00 | ||
|
Methylation in Case |
9.60E-02 (Median) | Methylation in Control | 1.22E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg04569615) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.33E+00 | Statistic Test | p-value:2.23E-08; Z-score:2.77E+00 | ||
|
Methylation in Case |
6.48E-01 (Median) | Methylation in Control | 4.86E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg05572763) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.29E+00 | Statistic Test | p-value:8.61E-06; Z-score:-1.11E+00 | ||
|
Methylation in Case |
1.14E-01 (Median) | Methylation in Control | 1.47E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg20632573) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.24E+00 | Statistic Test | p-value:1.88E-05; Z-score:-1.12E+00 | ||
|
Methylation in Case |
9.46E-02 (Median) | Methylation in Control | 1.17E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg17424999) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.35E+00 | Statistic Test | p-value:5.74E-05; Z-score:1.01E+00 | ||
|
Methylation in Case |
1.18E-01 (Median) | Methylation in Control | 8.75E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg05399320) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:1.49E-04; Z-score:-1.04E+00 | ||
|
Methylation in Case |
4.83E-01 (Median) | Methylation in Control | 5.56E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg21591624) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:9.17E-04; Z-score:9.83E-01 | ||
|
Methylation in Case |
2.48E-01 (Median) | Methylation in Control | 2.22E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg18042724) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:9.38E-04; Z-score:4.40E-01 | ||
|
Methylation in Case |
1.14E-01 (Median) | Methylation in Control | 1.05E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg00007644) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:4.59E-03; Z-score:-2.09E-01 | ||
|
Methylation in Case |
1.37E-01 (Median) | Methylation in Control | 1.41E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg10841552) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:1.75E-02; Z-score:3.49E-01 | ||
|
Methylation in Case |
1.93E-01 (Median) | Methylation in Control | 1.82E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC6A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg04032066) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:4.86E-02; Z-score:-6.97E-03 | ||
|
Methylation in Case |
1.65E-01 (Median) | Methylation in Control | 1.65E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Pancretic ductal adenocarcinoma |
17 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC6A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
5'UTR (cg20227806) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.33E+00 | Statistic Test | p-value:1.52E-25; Z-score:6.11E+00 | ||
|
Methylation in Case |
2.20E-01 (Median) | Methylation in Control | 9.41E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC6A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
5'UTR (cg14313833) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:7.97E-15; Z-score:-2.16E+00 | ||
|
Methylation in Case |
7.69E-01 (Median) | Methylation in Control | 8.61E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC6A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
5'UTR (cg10478435) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:2.70E-06; Z-score:1.98E-01 | ||
|
Methylation in Case |
3.63E-02 (Median) | Methylation in Control | 3.38E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC6A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
TSS1500 (cg22083639) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.33E+00 | Statistic Test | p-value:1.99E-11; Z-score:1.39E+00 | ||
|
Methylation in Case |
1.01E-01 (Median) | Methylation in Control | 7.59E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC6A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
TSS1500 (cg02868222) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.38E+00 | Statistic Test | p-value:2.17E-07; Z-score:-1.35E+00 | ||
|
Methylation in Case |
2.03E-01 (Median) | Methylation in Control | 2.79E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC6A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
TSS1500 (cg04721098) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:1.74E-05; Z-score:-8.80E-01 | ||
|
Methylation in Case |
5.31E-01 (Median) | Methylation in Control | 5.81E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC6A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
TSS200 (cg12180703) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.51E+00 | Statistic Test | p-value:3.80E-10; Z-score:2.03E+00 | ||
|
Methylation in Case |
4.75E-01 (Median) | Methylation in Control | 1.89E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC6A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
TSS200 (cg26925717) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.23E+00 | Statistic Test | p-value:1.87E-08; Z-score:-1.12E+00 | ||
|
Methylation in Case |
3.67E-01 (Median) | Methylation in Control | 4.50E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC6A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
TSS200 (cg07581732) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.11E+00 | Statistic Test | p-value:1.44E-06; Z-score:1.69E+00 | ||
|
Methylation in Case |
3.22E-01 (Median) | Methylation in Control | 1.53E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC6A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
1stExon (cg14730445) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:4.42E+00 | Statistic Test | p-value:1.94E-09; Z-score:2.26E+00 | ||
|
Methylation in Case |
4.70E-01 (Median) | Methylation in Control | 1.06E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC6A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
Body (cg02906784) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.20E+00 | Statistic Test | p-value:6.78E-16; Z-score:-2.15E+00 | ||
|
Methylation in Case |
5.85E-01 (Median) | Methylation in Control | 7.03E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC6A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
Body (cg05808709) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.19E+00 | Statistic Test | p-value:1.19E-15; Z-score:-2.00E+00 | ||
|
Methylation in Case |
5.99E-01 (Median) | Methylation in Control | 7.11E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC6A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
Body (cg00087274) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:1.16E-14; Z-score:-2.90E+00 | ||
|
Methylation in Case |
7.16E-01 (Median) | Methylation in Control | 7.90E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC6A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
Body (cg01995743) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:4.15E+00 | Statistic Test | p-value:1.59E-12; Z-score:2.57E+00 | ||
|
Methylation in Case |
4.03E-01 (Median) | Methylation in Control | 9.71E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC6A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
Body (cg06121808) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.74E+00 | Statistic Test | p-value:8.19E-12; Z-score:-2.11E+00 | ||
|
Methylation in Case |
3.59E-01 (Median) | Methylation in Control | 6.25E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC6A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
Body (cg13162609) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.07E-04; Z-score:-2.00E-01 | ||
|
Methylation in Case |
8.48E-01 (Median) | Methylation in Control | 8.52E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC6A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
3'UTR (cg24302133) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:2.30E-05; Z-score:1.24E+00 | ||
|
Methylation in Case |
8.45E-01 (Median) | Methylation in Control | 7.37E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
34 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg27036111) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.03E+00 | Statistic Test | p-value:8.74E-10; Z-score:1.11E+01 | ||
|
Methylation in Case |
6.32E-01 (Median) | Methylation in Control | 3.10E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg04592728) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.95E+00 | Statistic Test | p-value:5.40E-07; Z-score:6.48E+00 | ||
|
Methylation in Case |
4.68E-01 (Median) | Methylation in Control | 2.40E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg11288764) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.84E+00 | Statistic Test | p-value:9.05E-07; Z-score:1.31E+01 | ||
|
Methylation in Case |
2.70E-01 (Median) | Methylation in Control | 9.51E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg03573068) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:3.72E+00 | Statistic Test | p-value:1.17E-06; Z-score:1.86E+01 | ||
|
Methylation in Case |
5.11E-01 (Median) | Methylation in Control | 1.38E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg24420041) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.16E+00 | Statistic Test | p-value:2.66E-06; Z-score:1.03E+01 | ||
|
Methylation in Case |
2.04E-01 (Median) | Methylation in Control | 9.45E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg09696301) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.79E+00 | Statistic Test | p-value:1.35E-05; Z-score:1.78E+01 | ||
|
Methylation in Case |
1.60E-01 (Median) | Methylation in Control | 5.73E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg11784785) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.17E+00 | Statistic Test | p-value:2.46E-05; Z-score:5.29E+00 | ||
|
Methylation in Case |
2.15E-01 (Median) | Methylation in Control | 9.86E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
TSS200 (cg20551181) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:3.89E+00 | Statistic Test | p-value:3.38E-08; Z-score:8.55E+00 | ||
|
Methylation in Case |
4.87E-01 (Median) | Methylation in Control | 1.25E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
TSS200 (cg16635352) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.47E+00 | Statistic Test | p-value:3.92E-07; Z-score:1.79E+01 | ||
|
Methylation in Case |
5.09E-01 (Median) | Methylation in Control | 2.06E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
TSS200 (cg08530317) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.32E+00 | Statistic Test | p-value:1.41E-04; Z-score:8.57E+00 | ||
|
Methylation in Case |
1.70E-01 (Median) | Methylation in Control | 7.30E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
TSS200 (cg04642741) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.72E+00 | Statistic Test | p-value:3.39E-04; Z-score:9.28E+00 | ||
|
Methylation in Case |
4.72E-01 (Median) | Methylation in Control | 2.74E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
TSS200 (cg06804921) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.92E+00 | Statistic Test | p-value:9.78E-04; Z-score:8.71E+00 | ||
|
Methylation in Case |
6.09E-02 (Median) | Methylation in Control | 3.16E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg14524936) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.92E+00 | Statistic Test | p-value:9.08E-10; Z-score:1.02E+01 | ||
|
Methylation in Case |
6.97E-01 (Median) | Methylation in Control | 2.39E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg04032066) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:3.30E+00 | Statistic Test | p-value:1.32E-08; Z-score:1.54E+01 | ||
|
Methylation in Case |
4.42E-01 (Median) | Methylation in Control | 1.34E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg10841552) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:3.02E+00 | Statistic Test | p-value:1.51E-08; Z-score:3.50E+01 | ||
|
Methylation in Case |
4.16E-01 (Median) | Methylation in Control | 1.38E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg15828364) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.44E+00 | Statistic Test | p-value:1.25E-07; Z-score:6.91E+00 | ||
|
Methylation in Case |
4.39E-01 (Median) | Methylation in Control | 1.80E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg17424999) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:3.94E+00 | Statistic Test | p-value:2.19E-06; Z-score:2.00E+01 | ||
|
Methylation in Case |
2.60E-01 (Median) | Methylation in Control | 6.61E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg09136245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.69E+00 | Statistic Test | p-value:2.38E-06; Z-score:-6.82E+00 | ||
|
Methylation in Case |
6.14E-02 (Median) | Methylation in Control | 1.65E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg21591624) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.04E+00 | Statistic Test | p-value:1.10E-05; Z-score:5.47E+00 | ||
|
Methylation in Case |
3.18E-01 (Median) | Methylation in Control | 1.56E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg18042724) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.91E+00 | Statistic Test | p-value:3.81E-05; Z-score:4.74E+00 | ||
|
Methylation in Case |
1.87E-01 (Median) | Methylation in Control | 9.77E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg00007644) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.79E+00 | Statistic Test | p-value:5.36E-05; Z-score:5.68E+00 | ||
|
Methylation in Case |
2.13E-01 (Median) | Methylation in Control | 1.20E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg05572763) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.82E+00 | Statistic Test | p-value:5.76E-05; Z-score:-3.86E+00 | ||
|
Methylation in Case |
9.85E-02 (Median) | Methylation in Control | 1.80E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg19172170) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.81E+00 | Statistic Test | p-value:6.93E-05; Z-score:5.25E+00 | ||
|
Methylation in Case |
4.09E-01 (Median) | Methylation in Control | 2.26E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg04569615) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.72E+00 | Statistic Test | p-value:8.08E-05; Z-score:4.26E+00 | ||
|
Methylation in Case |
7.70E-01 (Median) | Methylation in Control | 4.48E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg04968806) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.90E+00 | Statistic Test | p-value:3.65E-04; Z-score:4.66E+00 | ||
|
Methylation in Case |
4.29E-01 (Median) | Methylation in Control | 2.26E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg05399320) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.28E+00 | Statistic Test | p-value:7.88E-04; Z-score:4.43E+00 | ||
|
Methylation in Case |
5.28E-01 (Median) | Methylation in Control | 4.14E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg09357935) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.71E+00 | Statistic Test | p-value:1.53E-03; Z-score:2.03E+00 | ||
|
Methylation in Case |
1.24E-01 (Median) | Methylation in Control | 7.26E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg21957058) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.51E+00 | Statistic Test | p-value:3.18E-03; Z-score:1.96E+00 | ||
|
Methylation in Case |
1.95E-01 (Median) | Methylation in Control | 1.29E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg12101354) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.42E+00 | Statistic Test | p-value:3.18E-03; Z-score:2.70E+00 | ||
|
Methylation in Case |
8.22E-02 (Median) | Methylation in Control | 5.77E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg20632573) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.36E+00 | Statistic Test | p-value:5.52E-03; Z-score:3.49E+00 | ||
|
Methylation in Case |
1.31E-01 (Median) | Methylation in Control | 9.59E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg23745839) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:6.93E-03; Z-score:1.21E-01 | ||
|
Methylation in Case |
1.45E-01 (Median) | Methylation in Control | 1.42E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg15083015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.56E+00 | Statistic Test | p-value:1.29E-02; Z-score:1.37E+00 | ||
|
Methylation in Case |
1.13E-01 (Median) | Methylation in Control | 7.22E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg03479705) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.34E+00 | Statistic Test | p-value:3.88E-02; Z-score:-5.86E+00 | ||
|
Methylation in Case |
2.55E-01 (Median) | Methylation in Control | 3.40E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC6A5 in bladder cancer | [ 4 ] | |||
|
Location |
3'UTR (cg08828816) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:4.69E-05; Z-score:-5.16E+00 | ||
|
Methylation in Case |
6.43E-01 (Median) | Methylation in Control | 7.60E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
35 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg27036111) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.35E+00 | Statistic Test | p-value:1.86E-12; Z-score:2.37E+00 | ||
|
Methylation in Case |
4.90E-01 (Median) | Methylation in Control | 3.64E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg04592728) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.46E+00 | Statistic Test | p-value:4.34E-11; Z-score:2.46E+00 | ||
|
Methylation in Case |
3.29E-01 (Median) | Methylation in Control | 2.25E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg03573068) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.75E+00 | Statistic Test | p-value:1.01E-10; Z-score:2.36E+00 | ||
|
Methylation in Case |
2.94E-01 (Median) | Methylation in Control | 1.69E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg09696301) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.83E+00 | Statistic Test | p-value:2.75E-09; Z-score:2.86E+00 | ||
|
Methylation in Case |
1.06E-01 (Median) | Methylation in Control | 5.79E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg11784785) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.50E+00 | Statistic Test | p-value:1.45E-08; Z-score:1.82E+00 | ||
|
Methylation in Case |
1.05E-01 (Median) | Methylation in Control | 7.00E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg11288764) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:2.59E-06; Z-score:8.79E-01 | ||
|
Methylation in Case |
1.38E-01 (Median) | Methylation in Control | 1.16E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg24420041) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:5.49E-05; Z-score:7.08E-01 | ||
|
Methylation in Case |
1.08E-01 (Median) | Methylation in Control | 8.87E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
TSS200 (cg16635352) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.03E+00 | Statistic Test | p-value:4.74E-22; Z-score:4.50E+00 | ||
|
Methylation in Case |
4.58E-01 (Median) | Methylation in Control | 2.26E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
TSS200 (cg20551181) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:3.76E+00 | Statistic Test | p-value:2.76E-21; Z-score:5.50E+00 | ||
|
Methylation in Case |
3.58E-01 (Median) | Methylation in Control | 9.52E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
TSS200 (cg04642741) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.30E+00 | Statistic Test | p-value:1.20E-07; Z-score:1.50E+00 | ||
|
Methylation in Case |
3.47E-01 (Median) | Methylation in Control | 2.67E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
TSS200 (cg06804921) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.85E+00 | Statistic Test | p-value:5.64E-06; Z-score:1.02E+00 | ||
|
Methylation in Case |
6.21E-02 (Median) | Methylation in Control | 3.35E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
TSS200 (cg08530317) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.33E+00 | Statistic Test | p-value:9.91E-06; Z-score:6.16E-01 | ||
|
Methylation in Case |
1.02E-01 (Median) | Methylation in Control | 7.65E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg04569615) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.48E+00 | Statistic Test | p-value:1.76E-18; Z-score:2.59E+00 | ||
|
Methylation in Case |
6.04E-01 (Median) | Methylation in Control | 4.09E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg21591624) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.47E+00 | Statistic Test | p-value:1.24E-14; Z-score:2.42E+00 | ||
|
Methylation in Case |
2.87E-01 (Median) | Methylation in Control | 1.95E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg15828364) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.74E+00 | Statistic Test | p-value:3.64E-14; Z-score:3.56E+00 | ||
|
Methylation in Case |
2.93E-01 (Median) | Methylation in Control | 1.68E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg04032066) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.79E+00 | Statistic Test | p-value:1.69E-13; Z-score:3.65E+00 | ||
|
Methylation in Case |
2.29E-01 (Median) | Methylation in Control | 1.28E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg00007644) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.60E+00 | Statistic Test | p-value:2.41E-13; Z-score:2.60E+00 | ||
|
Methylation in Case |
2.11E-01 (Median) | Methylation in Control | 1.32E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg10841552) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.68E+00 | Statistic Test | p-value:4.50E-12; Z-score:2.67E+00 | ||
|
Methylation in Case |
2.44E-01 (Median) | Methylation in Control | 1.45E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg14524936) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.74E+00 | Statistic Test | p-value:1.36E-11; Z-score:1.81E+00 | ||
|
Methylation in Case |
4.72E-01 (Median) | Methylation in Control | 2.71E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg17424999) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.13E+00 | Statistic Test | p-value:1.34E-10; Z-score:3.27E+00 | ||
|
Methylation in Case |
1.13E-01 (Median) | Methylation in Control | 5.33E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg18042724) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.84E+00 | Statistic Test | p-value:2.30E-10; Z-score:2.13E+00 | ||
|
Methylation in Case |
1.62E-01 (Median) | Methylation in Control | 8.81E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg09357935) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.77E+00 | Statistic Test | p-value:1.45E-09; Z-score:1.75E+00 | ||
|
Methylation in Case |
9.94E-02 (Median) | Methylation in Control | 5.61E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg18954388) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.32E+00 | Statistic Test | p-value:4.23E-08; Z-score:2.12E+00 | ||
|
Methylation in Case |
2.18E-01 (Median) | Methylation in Control | 1.64E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg04968806) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.51E+00 | Statistic Test | p-value:4.52E-07; Z-score:1.33E+00 | ||
|
Methylation in Case |
3.49E-01 (Median) | Methylation in Control | 2.31E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg13661968) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.24E+00 | Statistic Test | p-value:4.96E-07; Z-score:1.28E+00 | ||
|
Methylation in Case |
1.29E-01 (Median) | Methylation in Control | 1.04E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg19172170) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:6.47E-07; Z-score:1.16E+00 | ||
|
Methylation in Case |
2.82E-01 (Median) | Methylation in Control | 2.31E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg21957058) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.71E+00 | Statistic Test | p-value:6.87E-07; Z-score:1.43E+00 | ||
|
Methylation in Case |
2.04E-01 (Median) | Methylation in Control | 1.20E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg03771731) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.99E-06; Z-score:-9.24E-01 | ||
|
Methylation in Case |
7.77E-01 (Median) | Methylation in Control | 8.10E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg20632573) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.28E+00 | Statistic Test | p-value:3.00E-06; Z-score:1.27E+00 | ||
|
Methylation in Case |
1.25E-01 (Median) | Methylation in Control | 9.78E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg12101354) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:2.04E-05; Z-score:6.54E-01 | ||
|
Methylation in Case |
7.76E-02 (Median) | Methylation in Control | 6.52E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg15083015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:1.50E-04; Z-score:3.73E-01 | ||
|
Methylation in Case |
9.93E-02 (Median) | Methylation in Control | 8.72E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg07428491) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:1.90E-04; Z-score:8.35E-01 | ||
|
Methylation in Case |
2.22E-01 (Median) | Methylation in Control | 1.85E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg03479705) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.19E+00 | Statistic Test | p-value:5.14E-04; Z-score:-1.41E+00 | ||
|
Methylation in Case |
3.54E-01 (Median) | Methylation in Control | 4.23E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg05399320) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.20E+00 | Statistic Test | p-value:5.99E-04; Z-score:-1.01E+00 | ||
|
Methylation in Case |
5.45E-01 (Median) | Methylation in Control | 6.54E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC6A5 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg23745839) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.26E+00 | Statistic Test | p-value:1.47E-03; Z-score:8.60E-01 | ||
|
Methylation in Case |
1.52E-01 (Median) | Methylation in Control | 1.21E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
20 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg03573068) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.09E+00 | Statistic Test | p-value:3.73E-07; Z-score:2.59E+00 | ||
|
Methylation in Case |
2.55E-01 (Median) | Methylation in Control | 1.22E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg04592728) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.65E+00 | Statistic Test | p-value:1.28E-06; Z-score:4.14E+00 | ||
|
Methylation in Case |
4.31E-01 (Median) | Methylation in Control | 2.61E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg27036111) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.26E+00 | Statistic Test | p-value:2.23E-04; Z-score:1.90E+00 | ||
|
Methylation in Case |
5.05E-01 (Median) | Methylation in Control | 4.01E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg11784785) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.59E+00 | Statistic Test | p-value:2.42E-04; Z-score:1.62E+00 | ||
|
Methylation in Case |
8.28E-02 (Median) | Methylation in Control | 5.21E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg09696301) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.45E+00 | Statistic Test | p-value:2.89E-03; Z-score:1.19E+00 | ||
|
Methylation in Case |
4.88E-02 (Median) | Methylation in Control | 3.36E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg11288764) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.27E+00 | Statistic Test | p-value:6.00E-03; Z-score:6.78E-01 | ||
|
Methylation in Case |
9.45E-02 (Median) | Methylation in Control | 7.46E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg24420041) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:1.08E-02; Z-score:7.52E-01 | ||
|
Methylation in Case |
6.69E-02 (Median) | Methylation in Control | 5.53E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS200 (cg08530317) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.35E+00 | Statistic Test | p-value:4.28E-04; Z-score:6.73E-01 | ||
|
Methylation in Case |
4.83E-02 (Median) | Methylation in Control | 3.58E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS200 (cg06804921) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:1.69E-02; Z-score:1.01E+00 | ||
|
Methylation in Case |
3.03E-02 (Median) | Methylation in Control | 2.52E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg04569615) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.81E+00 | Statistic Test | p-value:2.88E-17; Z-score:3.86E+00 | ||
|
Methylation in Case |
7.04E-01 (Median) | Methylation in Control | 3.88E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg21591624) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.01E+00 | Statistic Test | p-value:1.47E-09; Z-score:3.01E+00 | ||
|
Methylation in Case |
2.48E-01 (Median) | Methylation in Control | 1.23E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg10841552) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.08E+00 | Statistic Test | p-value:3.85E-08; Z-score:3.04E+00 | ||
|
Methylation in Case |
2.05E-01 (Median) | Methylation in Control | 9.82E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg18954388) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.41E+00 | Statistic Test | p-value:4.11E-08; Z-score:2.01E+00 | ||
|
Methylation in Case |
1.92E-01 (Median) | Methylation in Control | 1.36E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg04032066) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.64E+00 | Statistic Test | p-value:9.93E-08; Z-score:4.16E+00 | ||
|
Methylation in Case |
1.86E-01 (Median) | Methylation in Control | 7.05E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg17424999) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.61E+00 | Statistic Test | p-value:1.70E-06; Z-score:3.46E+00 | ||
|
Methylation in Case |
7.89E-02 (Median) | Methylation in Control | 3.02E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg20632573) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.40E+00 | Statistic Test | p-value:9.89E-06; Z-score:1.33E+00 | ||
|
Methylation in Case |
8.34E-02 (Median) | Methylation in Control | 5.94E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg13661968) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.42E+00 | Statistic Test | p-value:3.00E-05; Z-score:1.26E+00 | ||
|
Methylation in Case |
7.00E-02 (Median) | Methylation in Control | 4.94E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg00007644) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.38E+00 | Statistic Test | p-value:3.43E-05; Z-score:1.70E+00 | ||
|
Methylation in Case |
1.12E-01 (Median) | Methylation in Control | 8.14E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg12101354) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.38E+00 | Statistic Test | p-value:2.59E-03; Z-score:9.79E-01 | ||
|
Methylation in Case |
3.52E-02 (Median) | Methylation in Control | 2.56E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC6A5 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg26213155) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:9.69E-03; Z-score:9.00E-01 | ||
|
Methylation in Case |
5.75E-01 (Median) | Methylation in Control | 5.30E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
30 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg03573068) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.55E+00 | Statistic Test | p-value:1.76E-11; Z-score:2.91E+00 | ||
|
Methylation in Case |
6.88E-01 (Median) | Methylation in Control | 4.43E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg24420041) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.73E+00 | Statistic Test | p-value:2.51E-11; Z-score:3.26E+00 | ||
|
Methylation in Case |
4.46E-01 (Median) | Methylation in Control | 2.59E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg11288764) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.99E+00 | Statistic Test | p-value:4.25E-11; Z-score:3.77E+00 | ||
|
Methylation in Case |
5.18E-01 (Median) | Methylation in Control | 2.61E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg09696301) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.73E+00 | Statistic Test | p-value:6.80E-09; Z-score:2.42E+00 | ||
|
Methylation in Case |
4.44E-01 (Median) | Methylation in Control | 2.57E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg04592728) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.35E+00 | Statistic Test | p-value:7.97E-07; Z-score:2.67E+00 | ||
|
Methylation in Case |
6.74E-01 (Median) | Methylation in Control | 5.00E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg11784785) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.37E+00 | Statistic Test | p-value:4.97E-05; Z-score:1.55E+00 | ||
|
Methylation in Case |
4.14E-01 (Median) | Methylation in Control | 3.02E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg27036111) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:3.45E-04; Z-score:1.13E+00 | ||
|
Methylation in Case |
7.99E-01 (Median) | Methylation in Control | 6.95E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS200 (cg08530317) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.74E+00 | Statistic Test | p-value:9.44E-07; Z-score:1.66E+00 | ||
|
Methylation in Case |
3.26E-01 (Median) | Methylation in Control | 1.87E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS200 (cg20551181) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.58E+00 | Statistic Test | p-value:9.80E-06; Z-score:1.41E+00 | ||
|
Methylation in Case |
4.94E-01 (Median) | Methylation in Control | 3.13E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS200 (cg04642741) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.25E+00 | Statistic Test | p-value:2.02E-05; Z-score:1.72E+00 | ||
|
Methylation in Case |
7.28E-01 (Median) | Methylation in Control | 5.80E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS200 (cg16635352) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.23E+00 | Statistic Test | p-value:5.28E-04; Z-score:1.29E+00 | ||
|
Methylation in Case |
8.01E-01 (Median) | Methylation in Control | 6.53E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS200 (cg06804921) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:4.23E-02; Z-score:3.82E-01 | ||
|
Methylation in Case |
2.15E-01 (Median) | Methylation in Control | 1.78E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg17424999) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.51E+00 | Statistic Test | p-value:1.65E-14; Z-score:3.25E+00 | ||
|
Methylation in Case |
4.09E-01 (Median) | Methylation in Control | 1.63E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg00007644) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.59E+00 | Statistic Test | p-value:2.03E-11; Z-score:2.49E+00 | ||
|
Methylation in Case |
5.99E-01 (Median) | Methylation in Control | 3.76E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg10841552) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.42E+00 | Statistic Test | p-value:1.21E-08; Z-score:1.83E+00 | ||
|
Methylation in Case |
6.68E-01 (Median) | Methylation in Control | 4.69E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg15828364) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.39E+00 | Statistic Test | p-value:6.35E-07; Z-score:1.66E+00 | ||
|
Methylation in Case |
6.57E-01 (Median) | Methylation in Control | 4.74E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg03479705) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:7.39E-07; Z-score:-1.46E+00 | ||
|
Methylation in Case |
5.36E-01 (Median) | Methylation in Control | 6.16E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg21957058) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.40E+00 | Statistic Test | p-value:9.03E-07; Z-score:1.62E+00 | ||
|
Methylation in Case |
4.60E-01 (Median) | Methylation in Control | 3.28E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg04032066) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.39E+00 | Statistic Test | p-value:1.40E-06; Z-score:1.59E+00 | ||
|
Methylation in Case |
7.17E-01 (Median) | Methylation in Control | 5.17E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg03771731) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:2.17E-06; Z-score:-3.17E+00 | ||
|
Methylation in Case |
8.54E-01 (Median) | Methylation in Control | 9.09E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg21591624) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.28E+00 | Statistic Test | p-value:9.44E-06; Z-score:1.43E+00 | ||
|
Methylation in Case |
6.31E-01 (Median) | Methylation in Control | 4.94E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg09357935) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.66E+00 | Statistic Test | p-value:1.08E-05; Z-score:1.43E+00 | ||
|
Methylation in Case |
2.44E-01 (Median) | Methylation in Control | 1.47E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg14524936) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:1.70E-04; Z-score:1.19E+00 | ||
|
Methylation in Case |
7.84E-01 (Median) | Methylation in Control | 6.47E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg04968806) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.23E+00 | Statistic Test | p-value:6.23E-04; Z-score:1.13E+00 | ||
|
Methylation in Case |
4.83E-01 (Median) | Methylation in Control | 3.93E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg04569615) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:3.49E-03; Z-score:1.13E+00 | ||
|
Methylation in Case |
8.41E-01 (Median) | Methylation in Control | 7.62E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg09136245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.33E+00 | Statistic Test | p-value:4.56E-03; Z-score:-1.24E+00 | ||
|
Methylation in Case |
1.75E-01 (Median) | Methylation in Control | 2.33E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg23745839) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:7.39E-03; Z-score:7.31E-01 | ||
|
Methylation in Case |
4.07E-01 (Median) | Methylation in Control | 3.42E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg26213155) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:1.11E-02; Z-score:7.27E-01 | ||
|
Methylation in Case |
5.89E-01 (Median) | Methylation in Control | 5.45E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC6A5 in colorectal cancer | [ 7 ] | |||
|
Location |
3'UTR (cg08828816) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:7.10E-07; Z-score:-3.11E+00 | ||
|
Methylation in Case |
8.16E-01 (Median) | Methylation in Control | 8.72E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
HIV infection |
30 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
TSS1500 (cg27036111) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.25E+00 | Statistic Test | p-value:3.08E-04; Z-score:1.25E+00 | ||
|
Methylation in Case |
4.72E-01 (Median) | Methylation in Control | 3.78E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
TSS1500 (cg04592728) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.16E+00 | Statistic Test | p-value:9.17E-04; Z-score:9.60E-01 | ||
|
Methylation in Case |
4.53E-01 (Median) | Methylation in Control | 3.90E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
TSS1500 (cg09696301) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.24E+00 | Statistic Test | p-value:1.91E-03; Z-score:1.02E+00 | ||
|
Methylation in Case |
1.34E-01 (Median) | Methylation in Control | 1.09E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
TSS1500 (cg24420041) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.26E+00 | Statistic Test | p-value:3.82E-03; Z-score:8.91E-01 | ||
|
Methylation in Case |
1.16E-01 (Median) | Methylation in Control | 9.19E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
TSS1500 (cg11784785) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.42E+00 | Statistic Test | p-value:4.85E-03; Z-score:1.15E+00 | ||
|
Methylation in Case |
1.56E-01 (Median) | Methylation in Control | 1.10E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
TSS1500 (cg03573068) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:1.22E-02; Z-score:5.11E-01 | ||
|
Methylation in Case |
2.24E-01 (Median) | Methylation in Control | 1.88E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
TSS200 (cg20551181) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.59E+00 | Statistic Test | p-value:3.31E-04; Z-score:1.28E+00 | ||
|
Methylation in Case |
2.23E-01 (Median) | Methylation in Control | 1.40E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
TSS200 (cg06804921) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.70E+00 | Statistic Test | p-value:2.14E-03; Z-score:1.05E+00 | ||
|
Methylation in Case |
5.68E-02 (Median) | Methylation in Control | 3.35E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
TSS200 (cg08530317) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.36E+00 | Statistic Test | p-value:2.54E-03; Z-score:1.06E+00 | ||
|
Methylation in Case |
8.93E-02 (Median) | Methylation in Control | 6.58E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
TSS200 (cg16635352) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:1.07E-02; Z-score:8.95E-01 | ||
|
Methylation in Case |
3.12E-01 (Median) | Methylation in Control | 2.56E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg05572763) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.42E+00 | Statistic Test | p-value:3.15E-08; Z-score:2.52E+00 | ||
|
Methylation in Case |
1.65E-01 (Median) | Methylation in Control | 1.17E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg18954388) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.26E+00 | Statistic Test | p-value:1.70E-06; Z-score:1.76E+00 | ||
|
Methylation in Case |
2.57E-01 (Median) | Methylation in Control | 2.03E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg17424999) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.89E+00 | Statistic Test | p-value:1.97E-06; Z-score:2.92E+00 | ||
|
Methylation in Case |
1.24E-01 (Median) | Methylation in Control | 6.59E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg04968806) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:2.27E-06; Z-score:1.70E+00 | ||
|
Methylation in Case |
3.81E-01 (Median) | Methylation in Control | 3.18E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg04032066) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.42E+00 | Statistic Test | p-value:2.91E-06; Z-score:2.35E+00 | ||
|
Methylation in Case |
2.12E-01 (Median) | Methylation in Control | 1.49E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg10841552) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.27E+00 | Statistic Test | p-value:2.63E-05; Z-score:1.33E+00 | ||
|
Methylation in Case |
2.18E-01 (Median) | Methylation in Control | 1.71E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg14524936) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.26E+00 | Statistic Test | p-value:4.69E-05; Z-score:1.19E+00 | ||
|
Methylation in Case |
4.57E-01 (Median) | Methylation in Control | 3.64E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg09136245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.33E+00 | Statistic Test | p-value:2.04E-04; Z-score:1.14E+00 | ||
|
Methylation in Case |
1.58E-01 (Median) | Methylation in Control | 1.19E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg20632573) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.27E+00 | Statistic Test | p-value:2.17E-04; Z-score:1.92E+00 | ||
|
Methylation in Case |
1.52E-01 (Median) | Methylation in Control | 1.20E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg21591624) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.18E+00 | Statistic Test | p-value:2.88E-04; Z-score:1.10E+00 | ||
|
Methylation in Case |
2.42E-01 (Median) | Methylation in Control | 2.05E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg26213155) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.16E+00 | Statistic Test | p-value:2.93E-04; Z-score:1.14E+00 | ||
|
Methylation in Case |
5.86E-01 (Median) | Methylation in Control | 5.06E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg04569615) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:2.01E-03; Z-score:1.12E+00 | ||
|
Methylation in Case |
6.17E-01 (Median) | Methylation in Control | 5.08E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg18042724) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.42E+00 | Statistic Test | p-value:3.01E-03; Z-score:9.74E-01 | ||
|
Methylation in Case |
1.12E-01 (Median) | Methylation in Control | 7.88E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg18855426) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.31E+00 | Statistic Test | p-value:8.91E-03; Z-score:6.69E-01 | ||
|
Methylation in Case |
3.84E-02 (Median) | Methylation in Control | 2.94E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg00007644) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:9.69E-03; Z-score:8.45E-01 | ||
|
Methylation in Case |
1.51E-01 (Median) | Methylation in Control | 1.26E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg13661968) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:1.03E-02; Z-score:8.17E-01 | ||
|
Methylation in Case |
2.05E-01 (Median) | Methylation in Control | 1.75E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg09357935) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.43E+00 | Statistic Test | p-value:2.34E-02; Z-score:1.01E+00 | ||
|
Methylation in Case |
1.11E-01 (Median) | Methylation in Control | 7.78E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg15828364) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.29E+00 | Statistic Test | p-value:3.23E-02; Z-score:8.94E-01 | ||
|
Methylation in Case |
2.43E-01 (Median) | Methylation in Control | 1.88E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg19172170) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:3.51E-02; Z-score:7.88E-01 | ||
|
Methylation in Case |
3.46E-01 (Median) | Methylation in Control | 2.88E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC6A5 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg21957058) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.24E+00 | Statistic Test | p-value:3.75E-02; Z-score:6.46E-01 | ||
|
Methylation in Case |
2.03E-01 (Median) | Methylation in Control | 1.65E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
16 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC6A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
TSS1500 (cg04592728) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.36E+00 | Statistic Test | p-value:1.10E-02; Z-score:2.57E+00 | ||
|
Methylation in Case |
4.36E-01 (Median) | Methylation in Control | 3.21E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC6A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
TSS1500 (cg24420041) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.26E+00 | Statistic Test | p-value:1.48E-02; Z-score:1.90E+00 | ||
|
Methylation in Case |
1.70E-01 (Median) | Methylation in Control | 1.35E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC6A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
TSS1500 (cg09696301) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.58E+00 | Statistic Test | p-value:2.87E-02; Z-score:5.95E+00 | ||
|
Methylation in Case |
1.68E-01 (Median) | Methylation in Control | 1.07E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC6A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
TSS200 (cg20551181) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.60E+00 | Statistic Test | p-value:2.74E-03; Z-score:2.74E+00 | ||
|
Methylation in Case |
2.86E-01 (Median) | Methylation in Control | 1.79E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC6A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
TSS200 (cg08530317) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.70E+00 | Statistic Test | p-value:6.86E-03; Z-score:4.52E+00 | ||
|
Methylation in Case |
2.07E-01 (Median) | Methylation in Control | 1.22E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC6A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg10841552) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.95E+00 | Statistic Test | p-value:1.38E-03; Z-score:5.49E+00 | ||
|
Methylation in Case |
3.65E-01 (Median) | Methylation in Control | 1.88E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC6A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg04968806) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.52E+00 | Statistic Test | p-value:1.90E-03; Z-score:2.63E+00 | ||
|
Methylation in Case |
4.26E-01 (Median) | Methylation in Control | 2.80E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC6A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg00007644) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.84E+00 | Statistic Test | p-value:1.96E-03; Z-score:4.19E+00 | ||
|
Methylation in Case |
2.70E-01 (Median) | Methylation in Control | 1.47E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC6A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg21591624) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.64E+00 | Statistic Test | p-value:2.12E-03; Z-score:2.57E+00 | ||
|
Methylation in Case |
3.81E-01 (Median) | Methylation in Control | 2.32E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC6A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg12101354) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.29E+00 | Statistic Test | p-value:4.13E-03; Z-score:3.01E+00 | ||
|
Methylation in Case |
1.34E-01 (Median) | Methylation in Control | 1.04E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC6A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg04569615) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:7.36E-03; Z-score:1.20E+00 | ||
|
Methylation in Case |
5.94E-01 (Median) | Methylation in Control | 5.25E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC6A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg20632573) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.49E+00 | Statistic Test | p-value:8.37E-03; Z-score:4.64E+00 | ||
|
Methylation in Case |
2.13E-01 (Median) | Methylation in Control | 1.42E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC6A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg14524936) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.61E+00 | Statistic Test | p-value:1.23E-02; Z-score:2.76E+00 | ||
|
Methylation in Case |
5.60E-01 (Median) | Methylation in Control | 3.48E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC6A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg13661968) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.31E+00 | Statistic Test | p-value:3.57E-02; Z-score:1.52E+00 | ||
|
Methylation in Case |
2.90E-01 (Median) | Methylation in Control | 2.22E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC6A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg18954388) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.28E+00 | Statistic Test | p-value:4.43E-02; Z-score:1.91E+00 | ||
|
Methylation in Case |
3.28E-01 (Median) | Methylation in Control | 2.55E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC6A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg04032066) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.58E+00 | Statistic Test | p-value:4.45E-02; Z-score:1.32E+00 | ||
|
Methylation in Case |
3.39E-01 (Median) | Methylation in Control | 2.15E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC6A5 in panic disorder | [ 10 ] | |||
|
Location |
TSS1500 (cg03573068) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-9.39E-01 | Statistic Test | p-value:4.96E-02; Z-score:-3.56E-01 | ||
|
Methylation in Case |
-2.41E+00 (Median) | Methylation in Control | -2.26E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC6A5 in panic disorder | [ 10 ] | |||
|
Location |
Body (cg09136245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-9.62E-01 | Statistic Test | p-value:1.19E-03; Z-score:-3.35E-01 | ||
|
Methylation in Case |
-3.33E+00 (Median) | Methylation in Control | -3.21E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC6A5 in panic disorder | [ 10 ] | |||
|
Location |
Body (cg17424999) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-9.55E-01 | Statistic Test | p-value:1.09E-02; Z-score:-3.31E-01 | ||
|
Methylation in Case |
-4.79E+00 (Median) | Methylation in Control | -4.57E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC6A5 in panic disorder | [ 10 ] | |||
|
Location |
Body (cg18855426) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-9.88E-01 | Statistic Test | p-value:1.72E-02; Z-score:-2.11E-01 | ||
|
Methylation in Case |
-4.83E+00 (Median) | Methylation in Control | -4.77E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
19 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
TSS1500 (cg02272859) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:3.41E-03; Z-score:3.46E+00 | ||
|
Methylation in Case |
8.94E-01 (Median) | Methylation in Control | 8.11E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
TSS1500 (cg25670451) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:2.54E-02; Z-score:2.07E+00 | ||
|
Methylation in Case |
9.44E-01 (Median) | Methylation in Control | 9.20E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
TSS1500 (cg03886254) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:3.07E-02; Z-score:1.63E+00 | ||
|
Methylation in Case |
9.64E-01 (Median) | Methylation in Control | 9.45E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
TSS200 (cg07430237) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.34E+00 | Statistic Test | p-value:1.47E-02; Z-score:3.04E+00 | ||
|
Methylation in Case |
6.50E-02 (Median) | Methylation in Control | 4.84E-02 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
TSS200 (cg20440901) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.88E+00 | Statistic Test | p-value:1.82E-02; Z-score:3.81E+00 | ||
|
Methylation in Case |
9.34E-02 (Median) | Methylation in Control | 4.96E-02 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
TSS200 (cg14720455) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:3.41E-02; Z-score:1.74E+00 | ||
|
Methylation in Case |
8.68E-01 (Median) | Methylation in Control | 8.23E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
TSS200 (cg23722792) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:4.16E-02; Z-score:1.11E+00 | ||
|
Methylation in Case |
9.16E-01 (Median) | Methylation in Control | 8.77E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
Body (cg11577329) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.51E+00 | Statistic Test | p-value:3.16E-03; Z-score:-3.54E+00 | ||
|
Methylation in Case |
5.44E-01 (Median) | Methylation in Control | 8.23E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
Body (cg06702884) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:5.14E-03; Z-score:2.95E+00 | ||
|
Methylation in Case |
9.05E-01 (Median) | Methylation in Control | 8.45E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
Body (cg17969789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:9.17E-03; Z-score:2.19E+00 | ||
|
Methylation in Case |
8.52E-01 (Median) | Methylation in Control | 7.65E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
Body (cg24682551) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.76E+00 | Statistic Test | p-value:9.98E-03; Z-score:7.03E+00 | ||
|
Methylation in Case |
7.52E-01 (Median) | Methylation in Control | 4.27E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
Body (cg27304952) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.32E+00 | Statistic Test | p-value:1.15E-02; Z-score:-3.45E+00 | ||
|
Methylation in Case |
5.31E-01 (Median) | Methylation in Control | 7.02E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
Body (cg02079348) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:1.22E-02; Z-score:2.45E+00 | ||
|
Methylation in Case |
8.11E-01 (Median) | Methylation in Control | 7.17E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
Body (cg00652223) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:1.61E-02; Z-score:2.49E+00 | ||
|
Methylation in Case |
8.97E-01 (Median) | Methylation in Control | 8.02E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
Body (cg10029905) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:2.08E-02; Z-score:1.81E+00 | ||
|
Methylation in Case |
9.51E-01 (Median) | Methylation in Control | 9.14E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
Body (cg20438663) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:2.30E-02; Z-score:3.37E+00 | ||
|
Methylation in Case |
8.33E-01 (Median) | Methylation in Control | 7.31E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
Body (cg19931596) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.25E+00 | Statistic Test | p-value:2.37E-02; Z-score:-3.59E+00 | ||
|
Methylation in Case |
5.60E-01 (Median) | Methylation in Control | 7.02E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC6A5 in prostate cancer | [ 11 ] | |||
|
Location |
Body (cg09079613) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:4.87E-02; Z-score:2.74E+00 | ||
|
Methylation in Case |
7.42E-01 (Median) | Methylation in Control | 6.57E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Systemic lupus erythematosus |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC6A5 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
TSS1500 (cg11784785) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:1.64E-03; Z-score:-1.26E-01 | ||
|
Methylation in Case |
1.56E-01 (Median) | Methylation in Control | 1.63E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC6A5 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg04968806) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:1.17E-02; Z-score:3.14E-01 | ||
|
Methylation in Case |
3.75E-01 (Median) | Methylation in Control | 3.61E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC6A5 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg05399320) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:1.28E-02; Z-score:2.38E-01 | ||
|
Methylation in Case |
6.10E-01 (Median) | Methylation in Control | 5.94E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC6A5 in papillary thyroid cancer | [ 13 ] | |||
|
Location |
TSS200 (cg20551181) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.23E+00 | Statistic Test | p-value:4.25E-06; Z-score:9.75E-01 | ||
|
Methylation in Case |
1.92E-01 (Median) | Methylation in Control | 1.56E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC6A5 in papillary thyroid cancer | [ 13 ] | |||
|
Location |
TSS200 (cg06804921) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:3.28E-02; Z-score:-7.55E-02 | ||
|
Methylation in Case |
7.26E-02 (Median) | Methylation in Control | 7.39E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC6A5 in papillary thyroid cancer | [ 13 ] | |||
|
Location |
Body (cg18954388) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.18E+00 | Statistic Test | p-value:4.72E-07; Z-score:1.06E+00 | ||
|
Methylation in Case |
1.55E-01 (Median) | Methylation in Control | 1.31E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC6A5 in papillary thyroid cancer | [ 13 ] | |||
|
Location |
Body (cg21591624) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.30E+00 | Statistic Test | p-value:6.93E-06; Z-score:1.04E+00 | ||
|
Methylation in Case |
1.44E-01 (Median) | Methylation in Control | 1.10E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC6A5 in papillary thyroid cancer | [ 13 ] | |||
|
Location |
Body (cg03479705) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:6.54E-04; Z-score:-1.06E+00 | ||
|
Methylation in Case |
3.16E-01 (Median) | Methylation in Control | 3.72E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC6A5 in papillary thyroid cancer | [ 13 ] | |||
|
Location |
Body (cg10841552) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.18E+00 | Statistic Test | p-value:6.69E-04; Z-score:7.14E-01 | ||
|
Methylation in Case |
1.21E-01 (Median) | Methylation in Control | 1.03E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC6A5 in papillary thyroid cancer | [ 13 ] | |||
|
Location |
Body (cg20632573) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:1.47E-02; Z-score:4.65E-01 | ||
|
Methylation in Case |
8.55E-02 (Median) | Methylation in Control | 7.88E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC6A5 in papillary thyroid cancer | [ 13 ] | |||
|
Location |
Body (cg04032066) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:2.72E-02; Z-score:3.64E-01 | ||
|
Methylation in Case |
1.15E-01 (Median) | Methylation in Control | 1.05E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC6A5 in papillary thyroid cancer | [ 13 ] | |||
|
Location |
3'UTR (cg08828816) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:4.46E-16; Z-score:-2.78E+00 | ||
|
Methylation in Case |
6.96E-01 (Median) | Methylation in Control | 7.93E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Atypical teratoid rhabdoid tumor |
16 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC6A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg00007644) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.33E+00 | Statistic Test | p-value:1.42E-05; Z-score:1.08E+00 | ||
|
Methylation in Case |
7.89E-01 (Median) | Methylation in Control | 5.93E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC6A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg03479705) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.29E+00 | Statistic Test | p-value:3.50E-04; Z-score:1.12E+00 | ||
|
Methylation in Case |
7.04E-01 (Median) | Methylation in Control | 5.44E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC6A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg03771731) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:4.30E-04; Z-score:-9.79E-01 | ||
|
Methylation in Case |
7.52E-01 (Median) | Methylation in Control | 8.16E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC6A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg04032066) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.60E+00 | Statistic Test | p-value:5.38E-04; Z-score:-6.91E-01 | ||
|
Methylation in Case |
1.58E-01 (Median) | Methylation in Control | 2.53E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC6A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg04569615) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:8.76E-04; Z-score:-4.64E-01 | ||
|
Methylation in Case |
6.28E-01 (Median) | Methylation in Control | 6.76E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC6A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg04968806) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:1.07E-03; Z-score:4.18E-01 | ||
|
Methylation in Case |
8.78E-01 (Median) | Methylation in Control | 8.09E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC6A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg05399320) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:1.42E-03; Z-score:2.09E-01 | ||
|
Methylation in Case |
2.55E-01 (Median) | Methylation in Control | 2.40E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC6A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg05572763) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:1.55E-03; Z-score:4.47E-01 | ||
|
Methylation in Case |
7.57E-02 (Median) | Methylation in Control | 6.90E-02 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC6A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg07428491) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.23E+00 | Statistic Test | p-value:3.91E-03; Z-score:6.63E-01 | ||
|
Methylation in Case |
5.52E-01 (Median) | Methylation in Control | 4.49E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC6A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg09136245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:9.38E-03; Z-score:3.60E-01 | ||
|
Methylation in Case |
8.20E-01 (Median) | Methylation in Control | 7.77E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC6A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg09357935) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:1.02E-02; Z-score:-3.34E-01 | ||
|
Methylation in Case |
2.64E-01 (Median) | Methylation in Control | 3.10E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC6A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg10841552) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.84E-02; Z-score:-2.57E-01 | ||
|
Methylation in Case |
7.98E-01 (Median) | Methylation in Control | 8.14E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC6A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg12101354) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.67E-02; Z-score:-1.37E-01 | ||
|
Methylation in Case |
9.13E-01 (Median) | Methylation in Control | 9.20E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC6A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg13661968) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:3.76E-02; Z-score:2.08E-01 | ||
|
Methylation in Case |
9.04E-01 (Median) | Methylation in Control | 8.90E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC6A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
Body (cg14524936) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:4.71E-02; Z-score:-3.69E-01 | ||
|
Methylation in Case |
8.69E-01 (Median) | Methylation in Control | 9.17E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC6A5 in atypical teratoid rhabdoid tumor | [ 14 ] | |||
|
Location |
3'UTR (cg08828816) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.37E+00 | Statistic Test | p-value:7.23E-13; Z-score:1.89E+00 | ||
|
Methylation in Case |
7.93E-01 (Median) | Methylation in Control | 5.81E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Celiac disease |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC6A5 in celiac disease | [ 15 ] | |||
|
Location |
Body (cg18042724) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.55E+00 | Statistic Test | p-value:1.04E-02; Z-score:1.52E+00 | ||
|
Methylation in Case |
6.57E-02 (Median) | Methylation in Control | 4.23E-02 (Median) | ||
|
Studied Phenotype |
Celiac disease[ ICD-11:DA95] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Depression |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC6A5 in depression | [ 16 ] | |||
|
Location |
Body (cg14524936) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:2.56E-02; Z-score:-2.85E-01 | ||
|
Methylation in Case |
2.75E-01 (Median) | Methylation in Control | 2.95E-01 (Median) | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Oligodendroglial tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypermethylation of SLC6A5 in oligodendroglial tumour than that in healthy individual | ||||
Studied Phenotype |
Oligodendroglial tumour [ICD-11:2A00.Y1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:4.57E-05; Fold-change:0.222426906; Z-score:4.019954388 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
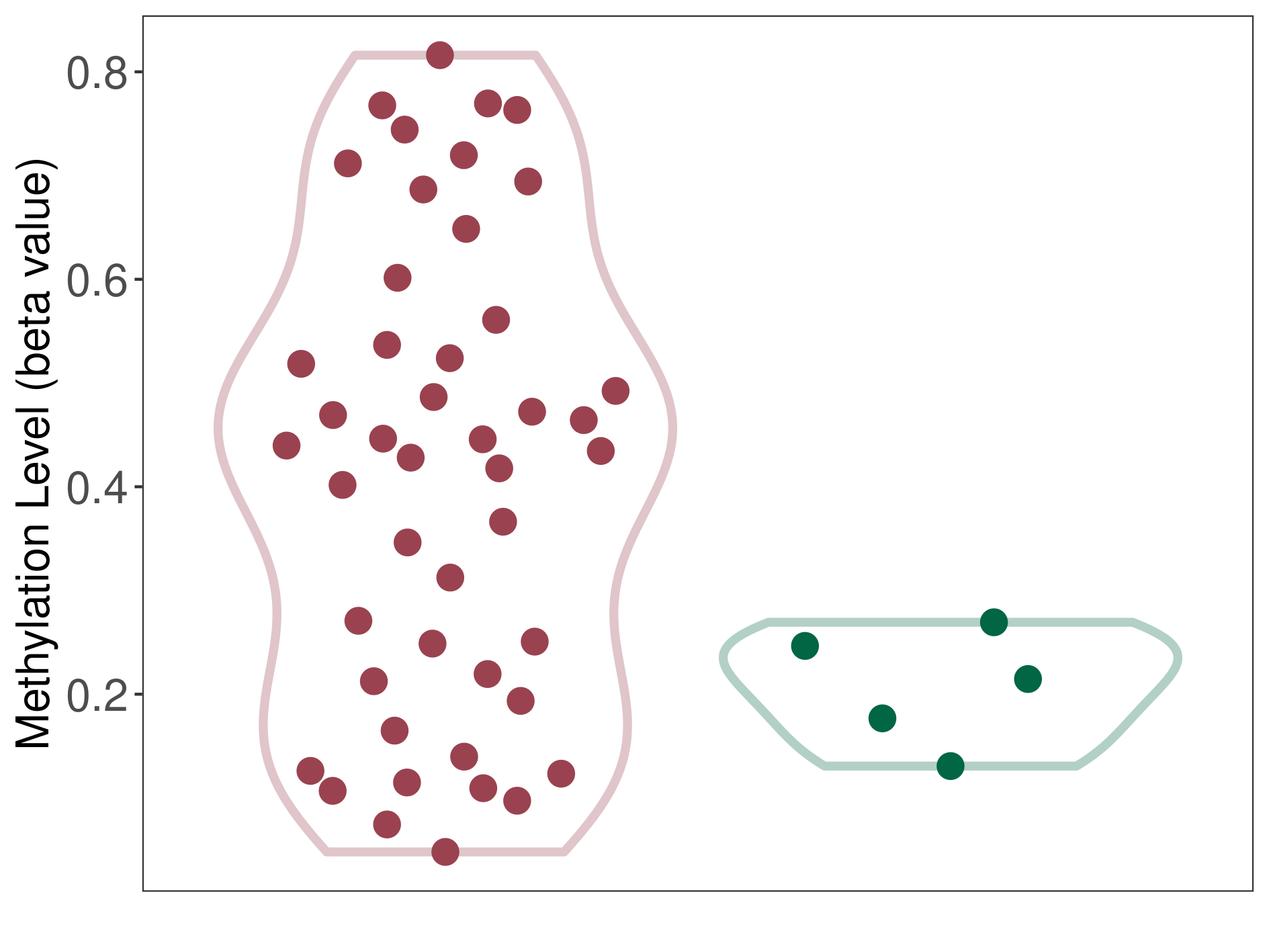
|
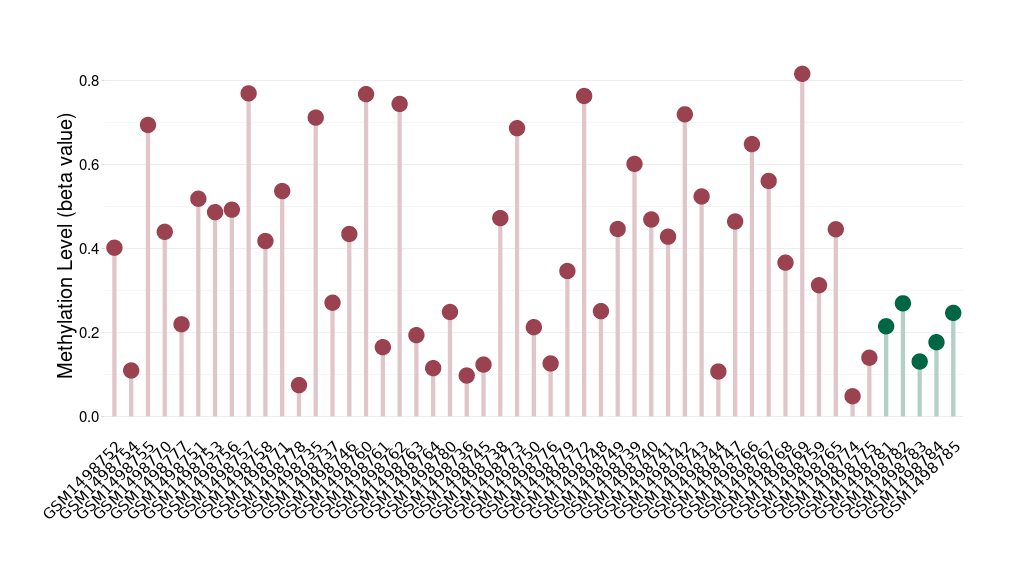 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Oligodendroglioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypermethylation of SLC6A5 in oligodendroglioma than that in healthy individual | ||||
Studied Phenotype |
Oligodendroglioma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:3.15E-20; Fold-change:0.249511318; Z-score:2.531204638 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
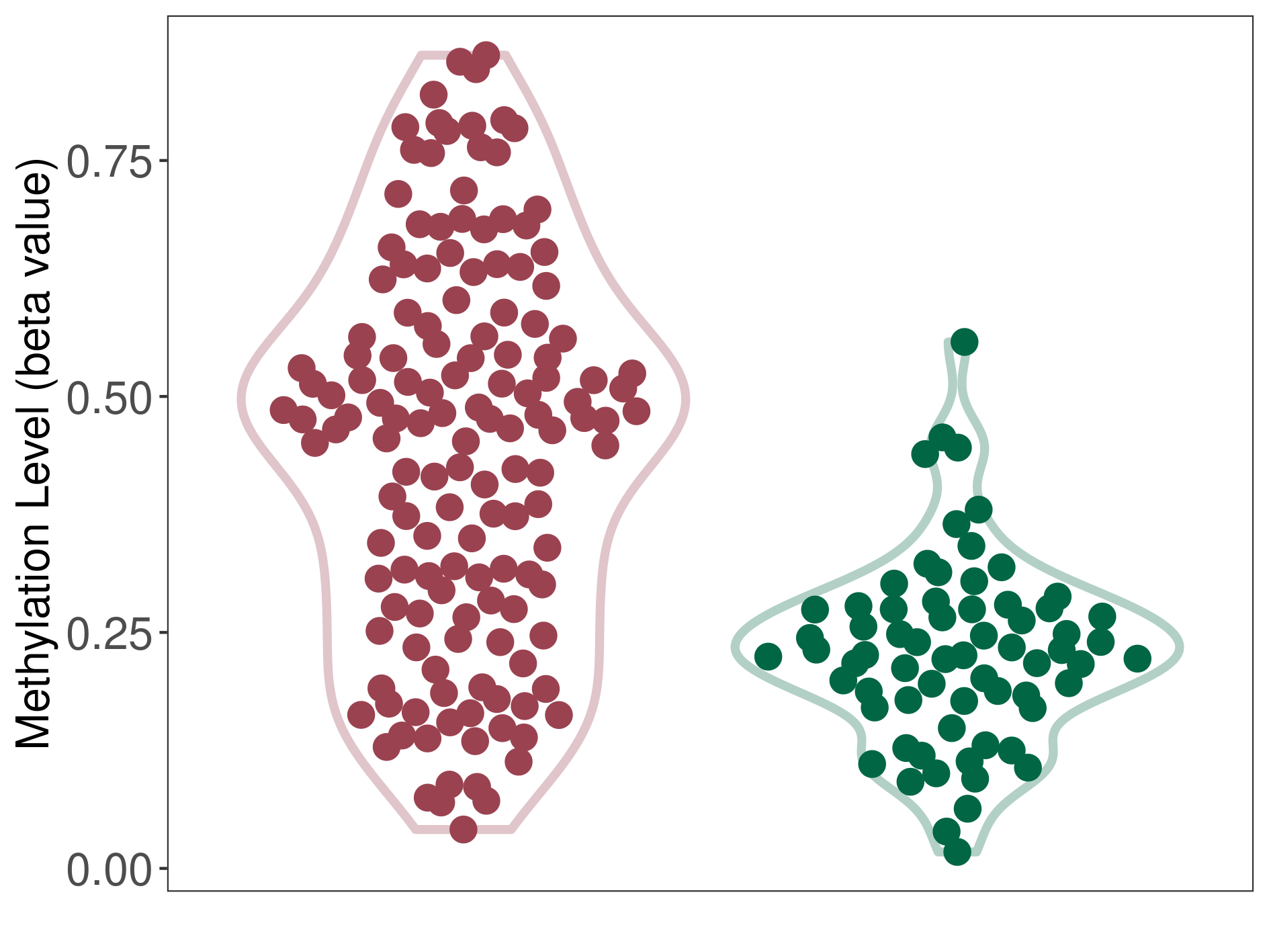
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Cerebellar liponeurocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC6A5 in cerebellar liponeurocytoma than that in healthy individual | ||||
Studied Phenotype |
Cerebellar liponeurocytoma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:6.81E-13; Fold-change:0.641114796; Z-score:8.56490826 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
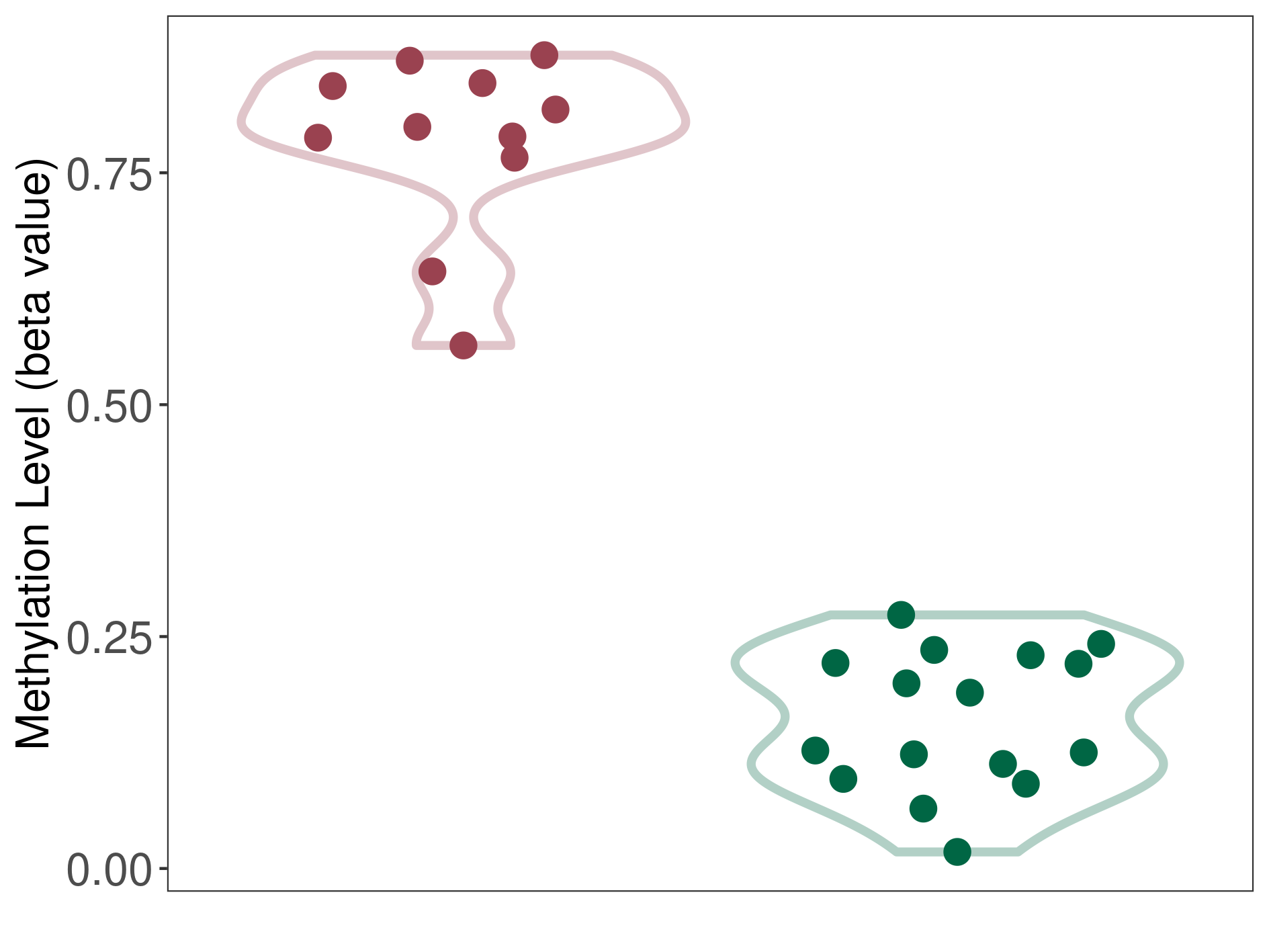
|
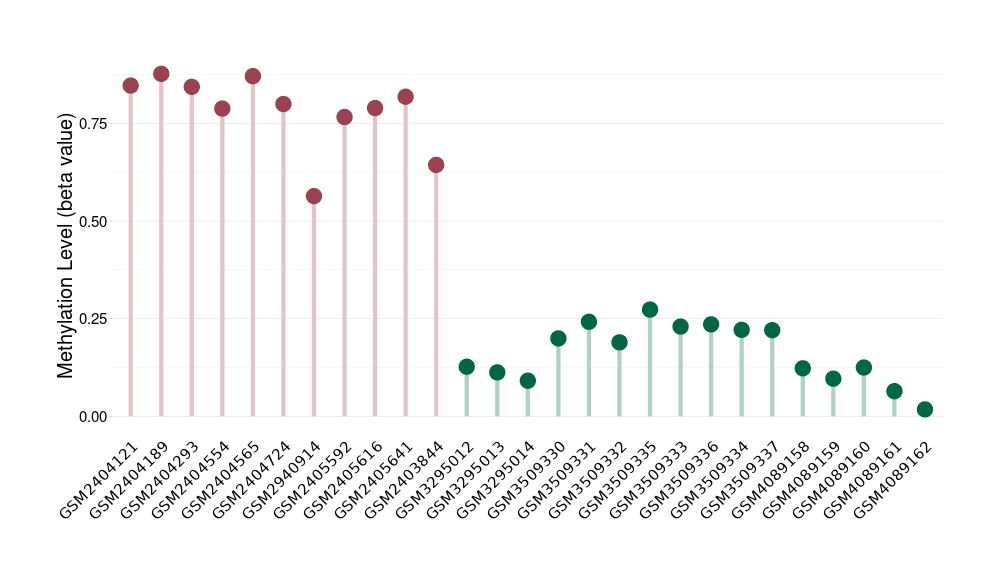 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Lymphoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC6A5 in lymphoma than that in healthy individual | ||||
Studied Phenotype |
Lymphoma [ICD-11:2B30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.91E-15; Fold-change:0.370809179; Z-score:3.757479342 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
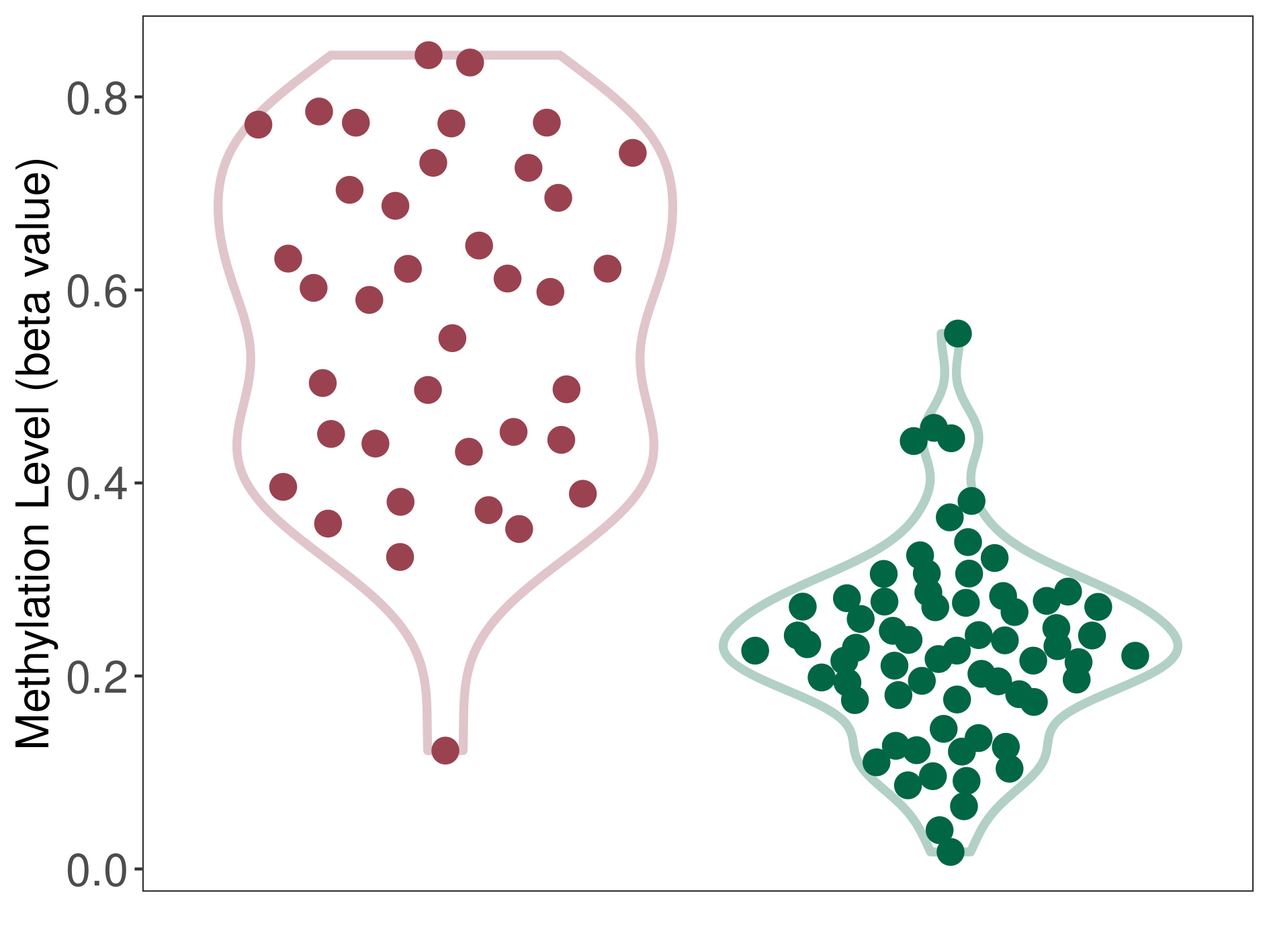
|
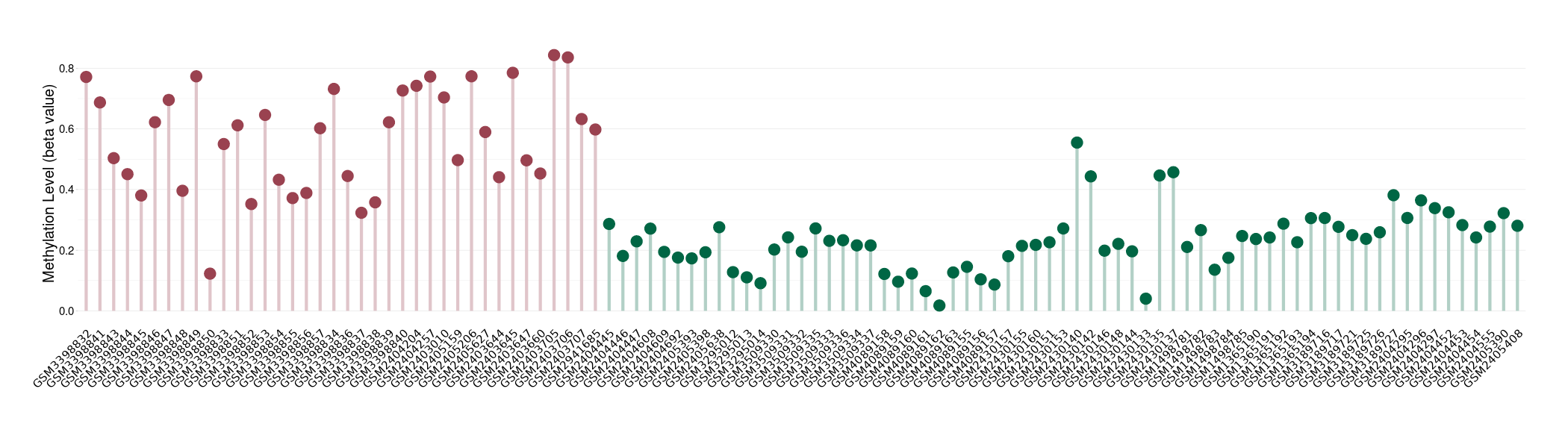 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
microRNA |
|||||
|
Unclear Phenotype |
12 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
miR-1272 directly targets SLC6A5 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-1272 | miRNA Mature ID | miR-1272 | ||
|
miRNA Sequence |
GAUGAUGAUGGCAGCAAAUUCUGAAA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon2 |
miR-1277 directly targets SLC6A5 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-1277 | miRNA Mature ID | miR-1277-5p | ||
|
miRNA Sequence |
AAAUAUAUAUAUAUAUGUACGUAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon3 |
miR-1322 directly targets SLC6A5 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-1322 | miRNA Mature ID | miR-1322 | ||
|
miRNA Sequence |
GAUGAUGCUGCUGAUGCUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon4 |
miR-190a directly targets SLC6A5 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-190a | miRNA Mature ID | miR-190a-3p | ||
|
miRNA Sequence |
CUAUAUAUCAAACAUAUUCCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon5 |
miR-194 directly targets SLC6A5 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-194 | miRNA Mature ID | miR-194-5p | ||
|
miRNA Sequence |
UGUAACAGCAACUCCAUGUGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon6 |
miR-4662a directly targets SLC6A5 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4662a | miRNA Mature ID | miR-4662a-3p | ||
|
miRNA Sequence |
AAAGAUAGACAAUUGGCUAAAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon7 |
miR-4711 directly targets SLC6A5 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4711 | miRNA Mature ID | miR-4711-5p | ||
|
miRNA Sequence |
UGCAUCAGGCCAGAAGACAUGAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon8 |
miR-4803 directly targets SLC6A5 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4803 | miRNA Mature ID | miR-4803 | ||
|
miRNA Sequence |
UAACAUAAUAGUGUGGAUUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon9 |
miR-5011 directly targets SLC6A5 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5011 | miRNA Mature ID | miR-5011-5p | ||
|
miRNA Sequence |
UAUAUAUACAGCCAUGCACUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon10 |
miR-579 directly targets SLC6A5 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-579 | miRNA Mature ID | miR-579-3p | ||
|
miRNA Sequence |
UUCAUUUGGUAUAAACCGCGAUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon11 |
miR-664b directly targets SLC6A5 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-664b | miRNA Mature ID | miR-664b-3p | ||
|
miRNA Sequence |
UUCAUUUGCCUCCCAGCCUACA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon12 |
miR-7112 directly targets SLC6A5 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-7112 | miRNA Mature ID | miR-7112-3p | ||
|
miRNA Sequence |
UGCAUCACAGCCUUUGGCCCUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
