Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0366 Transporter Info | ||||
| Gene Name | SLC44A4 | ||||
| Transporter Name | Choline transporter-like protein 4 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Pancretic ductal adenocarcinoma |
37 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg03427543) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:1.00E-11; Z-score:3.52E-01 | ||
|
Methylation in Case |
4.62E-02 (Median) | Methylation in Control | 4.33E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg21921016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.27E+00 | Statistic Test | p-value:1.86E-07; Z-score:1.41E+00 | ||
|
Methylation in Case |
7.14E-01 (Median) | Methylation in Control | 5.65E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg06578117) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:2.14E-07; Z-score:1.28E+00 | ||
|
Methylation in Case |
8.09E-01 (Median) | Methylation in Control | 6.90E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg08644341) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:7.97E-04; Z-score:-2.75E-01 | ||
|
Methylation in Case |
8.41E-01 (Median) | Methylation in Control | 8.48E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg20018106) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:6.84E-03; Z-score:7.94E-01 | ||
|
Methylation in Case |
7.94E-01 (Median) | Methylation in Control | 7.62E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg01939336) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.67E+00 | Statistic Test | p-value:1.22E-22; Z-score:4.23E+00 | ||
|
Methylation in Case |
1.84E-01 (Median) | Methylation in Control | 6.91E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg00791468) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:4.79E-10; Z-score:1.08E+00 | ||
|
Methylation in Case |
1.15E-01 (Median) | Methylation in Control | 9.59E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg23241456) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.48E+00 | Statistic Test | p-value:5.09E-09; Z-score:1.51E+00 | ||
|
Methylation in Case |
2.60E-01 (Median) | Methylation in Control | 1.75E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg04399565) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.45E+00 | Statistic Test | p-value:5.21E-07; Z-score:-1.64E+00 | ||
|
Methylation in Case |
5.03E-01 (Median) | Methylation in Control | 7.31E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg10015871) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:3.15E-05; Z-score:-1.64E+00 | ||
|
Methylation in Case |
2.70E-01 (Median) | Methylation in Control | 3.10E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg21692194) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:7.34E-05; Z-score:4.21E-01 | ||
|
Methylation in Case |
2.49E-01 (Median) | Methylation in Control | 2.25E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg08649013) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:4.00E-04; Z-score:9.88E-01 | ||
|
Methylation in Case |
8.56E-01 (Median) | Methylation in Control | 8.01E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg10189135) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:8.74E-04; Z-score:-5.32E-01 | ||
|
Methylation in Case |
4.42E-01 (Median) | Methylation in Control | 4.98E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg00825497) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:1.08E-03; Z-score:-7.56E-01 | ||
|
Methylation in Case |
9.06E-02 (Median) | Methylation in Control | 1.02E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg04074481) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:7.90E-03; Z-score:-2.30E-01 | ||
|
Methylation in Case |
7.47E-01 (Median) | Methylation in Control | 7.61E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg11466815) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:2.21E-02; Z-score:-3.46E-01 | ||
|
Methylation in Case |
4.03E-02 (Median) | Methylation in Control | 4.30E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg26685352) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.17E+00 | Statistic Test | p-value:6.03E-29; Z-score:5.07E+00 | ||
|
Methylation in Case |
2.65E-01 (Median) | Methylation in Control | 1.22E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg02509370) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.17E+00 | Statistic Test | p-value:5.18E-04; Z-score:-6.91E-01 | ||
|
Methylation in Case |
9.63E-02 (Median) | Methylation in Control | 1.13E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg13118536) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:3.69E-03; Z-score:-6.89E-01 | ||
|
Methylation in Case |
1.10E-01 (Median) | Methylation in Control | 1.23E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
1stExon (cg02318629) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.85E+00 | Statistic Test | p-value:1.01E-24; Z-score:3.85E+00 | ||
|
Methylation in Case |
3.58E-01 (Median) | Methylation in Control | 1.93E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
1stExon (cg04409954) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:2.83E-06; Z-score:-1.00E+00 | ||
|
Methylation in Case |
8.86E-01 (Median) | Methylation in Control | 9.08E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
1stExon (cg03040581) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:2.31E-02; Z-score:7.65E-01 | ||
|
Methylation in Case |
7.75E-01 (Median) | Methylation in Control | 7.37E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
1stExon (cg02192965) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:4.35E-02; Z-score:-4.75E-01 | ||
|
Methylation in Case |
5.82E-01 (Median) | Methylation in Control | 6.28E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg12111714) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.64E+00 | Statistic Test | p-value:1.61E-10; Z-score:2.20E+00 | ||
|
Methylation in Case |
7.14E-01 (Median) | Methylation in Control | 4.35E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg10718608) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:9.78E-10; Z-score:-1.84E+00 | ||
|
Methylation in Case |
5.52E-01 (Median) | Methylation in Control | 7.51E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg23503101) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:1.56E-08; Z-score:1.80E+00 | ||
|
Methylation in Case |
7.64E-01 (Median) | Methylation in Control | 6.97E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg21689824) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:1.78E-08; Z-score:-1.58E+00 | ||
|
Methylation in Case |
4.35E-01 (Median) | Methylation in Control | 5.00E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg23880589) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.26E+00 | Statistic Test | p-value:3.32E-08; Z-score:-2.06E+00 | ||
|
Methylation in Case |
2.77E-01 (Median) | Methylation in Control | 3.49E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg21961694) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.20E+00 | Statistic Test | p-value:1.60E-07; Z-score:-1.52E+00 | ||
|
Methylation in Case |
6.37E-01 (Median) | Methylation in Control | 7.62E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg07598082) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:1.93E-04; Z-score:6.34E-01 | ||
|
Methylation in Case |
8.35E-01 (Median) | Methylation in Control | 8.09E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg00981877) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.44E+00 | Statistic Test | p-value:7.30E-04; Z-score:-5.48E-01 | ||
|
Methylation in Case |
7.70E-02 (Median) | Methylation in Control | 1.11E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg00719108) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.00E+00 | Statistic Test | p-value:1.54E-03; Z-score:2.03E-01 | ||
|
Methylation in Case |
9.45E-01 (Median) | Methylation in Control | 9.43E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg01420001) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:1.75E-03; Z-score:-6.62E-01 | ||
|
Methylation in Case |
7.53E-01 (Median) | Methylation in Control | 7.88E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg07521929) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:1.86E-03; Z-score:-7.33E-01 | ||
|
Methylation in Case |
4.75E-01 (Median) | Methylation in Control | 5.10E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg17928607) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:3.14E-02; Z-score:-1.31E-01 | ||
|
Methylation in Case |
8.59E-01 (Median) | Methylation in Control | 8.62E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg14414100) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:4.42E-02; Z-score:-5.10E-01 | ||
|
Methylation in Case |
3.65E-01 (Median) | Methylation in Control | 3.95E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon37 |
Methylation of SLC44A4 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
3'UTR (cg11165756) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:1.30E-03; Z-score:7.77E-01 | ||
|
Methylation in Case |
8.94E-01 (Median) | Methylation in Control | 8.81E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
18 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
5'UTR (cg00997378) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:1.42E-02; Z-score:1.80E+00 | ||
|
Methylation in Case |
9.03E-01 (Median) | Methylation in Control | 8.25E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
5'UTR (cg02700824) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.80E+00 | Statistic Test | p-value:2.75E-02; Z-score:-1.90E+00 | ||
|
Methylation in Case |
1.74E-02 (Median) | Methylation in Control | 3.14E-02 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg19916794) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.26E+00 | Statistic Test | p-value:3.76E-03; Z-score:3.26E+00 | ||
|
Methylation in Case |
8.25E-01 (Median) | Methylation in Control | 6.55E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg21088438) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:2.83E-02; Z-score:1.67E+00 | ||
|
Methylation in Case |
8.84E-01 (Median) | Methylation in Control | 7.23E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg02659138) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.47E+00 | Statistic Test | p-value:4.10E-02; Z-score:-2.89E+00 | ||
|
Methylation in Case |
5.28E-01 (Median) | Methylation in Control | 7.77E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
TSS200 (cg16424326) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.48E+00 | Statistic Test | p-value:3.57E-02; Z-score:1.49E+00 | ||
|
Methylation in Case |
4.97E-02 (Median) | Methylation in Control | 3.36E-02 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
Body (cg03374976) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.73E+00 | Statistic Test | p-value:7.38E-03; Z-score:-7.76E+00 | ||
|
Methylation in Case |
3.63E-01 (Median) | Methylation in Control | 6.27E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
Body (cg02456521) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.25E+00 | Statistic Test | p-value:8.87E-03; Z-score:2.76E+00 | ||
|
Methylation in Case |
7.94E-01 (Median) | Methylation in Control | 6.37E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
Body (cg20282814) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.43E+00 | Statistic Test | p-value:1.48E-02; Z-score:-2.37E+00 | ||
|
Methylation in Case |
4.52E-01 (Median) | Methylation in Control | 6.47E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
Body (cg12486558) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:1.50E-02; Z-score:-1.14E+01 | ||
|
Methylation in Case |
6.76E-01 (Median) | Methylation in Control | 9.12E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
Body (cg16084788) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:2.33E-02; Z-score:1.49E+00 | ||
|
Methylation in Case |
9.47E-01 (Median) | Methylation in Control | 9.27E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
Body (cg02635677) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:2.41E-02; Z-score:2.37E+00 | ||
|
Methylation in Case |
8.53E-01 (Median) | Methylation in Control | 7.63E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
Body (cg04320476) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:2.42E-02; Z-score:3.49E+00 | ||
|
Methylation in Case |
9.44E-01 (Median) | Methylation in Control | 8.95E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
Body (cg06372223) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.49E+00 | Statistic Test | p-value:2.48E-02; Z-score:-2.36E+00 | ||
|
Methylation in Case |
3.48E-01 (Median) | Methylation in Control | 5.19E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
Body (cg14326305) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.46E+00 | Statistic Test | p-value:4.04E-02; Z-score:-2.03E+00 | ||
|
Methylation in Case |
2.95E-01 (Median) | Methylation in Control | 4.32E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
Body (cg03048902) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:4.31E-02; Z-score:1.89E+00 | ||
|
Methylation in Case |
8.96E-01 (Median) | Methylation in Control | 8.57E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
3'UTR (cg23253978) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.24E+00 | Statistic Test | p-value:7.20E-03; Z-score:2.97E+00 | ||
|
Methylation in Case |
6.15E-01 (Median) | Methylation in Control | 4.95E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC44A4 in prostate cancer | [ 2 ] | |||
|
Location |
3'UTR (cg19599386) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.41E+00 | Statistic Test | p-value:9.12E-03; Z-score:5.62E+00 | ||
|
Methylation in Case |
7.69E-01 (Median) | Methylation in Control | 5.47E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
53 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg16553272) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.24E+00 | Statistic Test | p-value:9.46E-05; Z-score:-9.23E+00 | ||
|
Methylation in Case |
4.44E-01 (Median) | Methylation in Control | 5.50E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
TSS200 (cg24529722) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:7.13E-06; Z-score:-5.75E+00 | ||
|
Methylation in Case |
3.52E-01 (Median) | Methylation in Control | 4.74E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
TSS200 (cg24707219) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.27E+00 | Statistic Test | p-value:1.33E-05; Z-score:-5.55E+00 | ||
|
Methylation in Case |
4.77E-01 (Median) | Methylation in Control | 6.05E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
TSS200 (cg04567302) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.28E+00 | Statistic Test | p-value:2.88E-05; Z-score:-4.00E+00 | ||
|
Methylation in Case |
4.22E-01 (Median) | Methylation in Control | 5.40E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
TSS200 (cg21236501) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.39E+00 | Statistic Test | p-value:3.98E-05; Z-score:-7.01E+00 | ||
|
Methylation in Case |
3.79E-01 (Median) | Methylation in Control | 5.26E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
TSS200 (cg03045620) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:2.13E-03; Z-score:-2.49E+00 | ||
|
Methylation in Case |
3.45E-01 (Median) | Methylation in Control | 3.96E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
TSS200 (cg07363637) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:1.47E-02; Z-score:-1.41E+00 | ||
|
Methylation in Case |
4.26E-01 (Median) | Methylation in Control | 4.68E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg07546508) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.59E+00 | Statistic Test | p-value:8.59E-11; Z-score:-1.17E+01 | ||
|
Methylation in Case |
4.43E-01 (Median) | Methylation in Control | 7.03E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg07185041) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.31E+00 | Statistic Test | p-value:8.35E-10; Z-score:-8.79E+00 | ||
|
Methylation in Case |
2.86E-01 (Median) | Methylation in Control | 6.61E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg01347702) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.36E+00 | Statistic Test | p-value:2.37E-09; Z-score:-7.98E+00 | ||
|
Methylation in Case |
2.57E-01 (Median) | Methylation in Control | 6.06E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg18856043) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.95E+00 | Statistic Test | p-value:2.07E-08; Z-score:-7.55E+00 | ||
|
Methylation in Case |
2.30E-01 (Median) | Methylation in Control | 4.50E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg27005847) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.81E+00 | Statistic Test | p-value:3.01E-08; Z-score:-6.90E+00 | ||
|
Methylation in Case |
3.67E-01 (Median) | Methylation in Control | 6.66E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg05686323) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.48E+00 | Statistic Test | p-value:7.37E-07; Z-score:-6.50E+00 | ||
|
Methylation in Case |
4.94E-01 (Median) | Methylation in Control | 7.31E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg24490633) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.67E+00 | Statistic Test | p-value:7.89E-07; Z-score:5.09E+00 | ||
|
Methylation in Case |
5.08E-01 (Median) | Methylation in Control | 3.03E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg07601645) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.41E+00 | Statistic Test | p-value:1.60E-06; Z-score:5.63E+00 | ||
|
Methylation in Case |
5.71E-01 (Median) | Methylation in Control | 4.06E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg11943056) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.35E+00 | Statistic Test | p-value:3.71E-06; Z-score:-5.92E+00 | ||
|
Methylation in Case |
2.81E-01 (Median) | Methylation in Control | 6.60E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg09130091) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.35E+00 | Statistic Test | p-value:4.04E-06; Z-score:4.59E+00 | ||
|
Methylation in Case |
4.63E-01 (Median) | Methylation in Control | 3.44E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg09153713) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.37E+00 | Statistic Test | p-value:6.98E-06; Z-score:-9.84E+00 | ||
|
Methylation in Case |
4.59E-01 (Median) | Methylation in Control | 6.28E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg23701033) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.31E+00 | Statistic Test | p-value:1.65E-05; Z-score:-7.93E+00 | ||
|
Methylation in Case |
4.48E-01 (Median) | Methylation in Control | 5.86E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg25013052) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.42E+00 | Statistic Test | p-value:1.77E-05; Z-score:4.44E+00 | ||
|
Methylation in Case |
7.26E-01 (Median) | Methylation in Control | 5.10E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg20961940) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:1.96E-05; Z-score:-9.18E+00 | ||
|
Methylation in Case |
4.74E-01 (Median) | Methylation in Control | 6.44E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg24732991) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:4.42E-05; Z-score:-6.75E+00 | ||
|
Methylation in Case |
7.22E-01 (Median) | Methylation in Control | 8.38E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg09298971) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.21E+00 | Statistic Test | p-value:6.06E-05; Z-score:-5.65E+00 | ||
|
Methylation in Case |
6.20E-01 (Median) | Methylation in Control | 7.53E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg20498033) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:7.59E-05; Z-score:-5.63E+00 | ||
|
Methylation in Case |
7.92E-01 (Median) | Methylation in Control | 8.48E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg19955284) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.30E+00 | Statistic Test | p-value:9.44E-05; Z-score:5.18E+00 | ||
|
Methylation in Case |
8.17E-01 (Median) | Methylation in Control | 6.27E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg04261164) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:1.54E-04; Z-score:3.10E+00 | ||
|
Methylation in Case |
8.51E-01 (Median) | Methylation in Control | 8.11E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg04021562) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.80E+00 | Statistic Test | p-value:1.80E-04; Z-score:-4.07E+00 | ||
|
Methylation in Case |
1.18E-01 (Median) | Methylation in Control | 2.11E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg27587935) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.41E+00 | Statistic Test | p-value:2.47E-04; Z-score:3.77E+00 | ||
|
Methylation in Case |
8.57E-01 (Median) | Methylation in Control | 6.07E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg07438099) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:3.32E-04; Z-score:2.69E+00 | ||
|
Methylation in Case |
6.78E-01 (Median) | Methylation in Control | 5.98E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg25368728) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.19E+00 | Statistic Test | p-value:4.28E-04; Z-score:-1.07E+01 | ||
|
Methylation in Case |
6.32E-01 (Median) | Methylation in Control | 7.55E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg18263014) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:4.91E-04; Z-score:-3.17E+00 | ||
|
Methylation in Case |
7.92E-01 (Median) | Methylation in Control | 8.41E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg07705820) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:5.07E-04; Z-score:-4.50E+00 | ||
|
Methylation in Case |
7.16E-01 (Median) | Methylation in Control | 7.89E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg02721751) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.32E+00 | Statistic Test | p-value:5.08E-04; Z-score:3.35E+00 | ||
|
Methylation in Case |
7.29E-01 (Median) | Methylation in Control | 5.51E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg22882121) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.26E+00 | Statistic Test | p-value:7.25E-04; Z-score:-3.31E+00 | ||
|
Methylation in Case |
5.02E-01 (Median) | Methylation in Control | 6.30E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg26434400) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:7.88E-04; Z-score:-1.93E+00 | ||
|
Methylation in Case |
8.60E-01 (Median) | Methylation in Control | 8.86E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg26472225) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:1.22E-03; Z-score:2.61E+00 | ||
|
Methylation in Case |
5.08E-01 (Median) | Methylation in Control | 4.27E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon37 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg26138846) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.47E+00 | Statistic Test | p-value:1.36E-03; Z-score:-5.35E+00 | ||
|
Methylation in Case |
4.10E-01 (Median) | Methylation in Control | 6.03E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon38 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg25600103) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.42E+00 | Statistic Test | p-value:1.66E-03; Z-score:-2.72E+00 | ||
|
Methylation in Case |
4.10E-01 (Median) | Methylation in Control | 5.83E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon39 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg20544437) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:2.24E-03; Z-score:3.00E+00 | ||
|
Methylation in Case |
8.41E-01 (Median) | Methylation in Control | 7.22E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon40 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg10951325) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.32E+00 | Statistic Test | p-value:3.23E-03; Z-score:-2.69E+00 | ||
|
Methylation in Case |
5.06E-01 (Median) | Methylation in Control | 6.66E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon41 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg04489104) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.37E+00 | Statistic Test | p-value:3.66E-03; Z-score:7.94E+00 | ||
|
Methylation in Case |
3.28E-01 (Median) | Methylation in Control | 2.40E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon42 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg02252769) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.27E+00 | Statistic Test | p-value:4.84E-03; Z-score:2.48E+00 | ||
|
Methylation in Case |
6.81E-01 (Median) | Methylation in Control | 5.38E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon43 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg19117698) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:5.29E-03; Z-score:3.25E+00 | ||
|
Methylation in Case |
7.50E-01 (Median) | Methylation in Control | 6.88E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon44 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg11312554) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:9.30E-03; Z-score:2.31E+00 | ||
|
Methylation in Case |
8.67E-01 (Median) | Methylation in Control | 8.30E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon45 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg00123181) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:1.14E-02; Z-score:1.89E+00 | ||
|
Methylation in Case |
7.99E-01 (Median) | Methylation in Control | 7.65E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon46 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg18949702) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.41E-02; Z-score:-3.09E+00 | ||
|
Methylation in Case |
7.54E-01 (Median) | Methylation in Control | 7.79E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon47 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg13975172) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:1.41E-02; Z-score:1.53E+00 | ||
|
Methylation in Case |
6.87E-01 (Median) | Methylation in Control | 5.78E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon48 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg22872508) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:1.55E-02; Z-score:1.67E+00 | ||
|
Methylation in Case |
8.60E-01 (Median) | Methylation in Control | 8.15E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon49 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg11640015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.32E+00 | Statistic Test | p-value:1.87E-02; Z-score:-1.90E+00 | ||
|
Methylation in Case |
4.95E-01 (Median) | Methylation in Control | 6.54E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon50 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg07205860) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:1.97E-02; Z-score:1.73E+00 | ||
|
Methylation in Case |
8.73E-01 (Median) | Methylation in Control | 8.33E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon51 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg24369466) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:2.12E-02; Z-score:-1.79E+00 | ||
|
Methylation in Case |
6.23E-01 (Median) | Methylation in Control | 6.88E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon52 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg10969178) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.38E+00 | Statistic Test | p-value:2.19E-02; Z-score:-1.68E+00 | ||
|
Methylation in Case |
5.22E-01 (Median) | Methylation in Control | 7.23E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon53 |
Methylation of SLC44A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg21618695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:4.01E-02; Z-score:1.91E+00 | ||
|
Methylation in Case |
8.24E-01 (Median) | Methylation in Control | 8.06E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colon cancer |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC44A4 in colon adenocarcinoma | [ 4 ] | |||
|
Location |
TSS1500 (cg20092728) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.17E+00 | Statistic Test | p-value:2.07E-03; Z-score:-3.66E+00 | ||
|
Methylation in Case |
6.79E-01 (Median) | Methylation in Control | 7.95E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC44A4 in colon adenocarcinoma | [ 4 ] | |||
|
Location |
Body (cg08382534) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.37E+00 | Statistic Test | p-value:6.21E-05; Z-score:-1.52E+00 | ||
|
Methylation in Case |
3.99E-01 (Median) | Methylation in Control | 5.49E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
HIV infection |
41 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
TSS1500 (cg16553272) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:2.34E-07; Z-score:1.11E+00 | ||
|
Methylation in Case |
8.55E-01 (Median) | Methylation in Control | 7.64E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
TSS200 (cg21236501) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.24E+00 | Statistic Test | p-value:8.35E-15; Z-score:2.65E+00 | ||
|
Methylation in Case |
6.32E-01 (Median) | Methylation in Control | 5.10E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
TSS200 (cg24529722) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.26E+00 | Statistic Test | p-value:4.43E-11; Z-score:2.67E+00 | ||
|
Methylation in Case |
6.26E-01 (Median) | Methylation in Control | 4.99E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
TSS200 (cg04567302) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:9.83E-11; Z-score:1.61E+00 | ||
|
Methylation in Case |
8.33E-01 (Median) | Methylation in Control | 7.22E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
TSS200 (cg03045620) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:3.39E-09; Z-score:2.15E+00 | ||
|
Methylation in Case |
6.13E-01 (Median) | Methylation in Control | 5.12E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
TSS200 (cg07363637) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:4.10E-09; Z-score:1.39E+00 | ||
|
Methylation in Case |
8.24E-01 (Median) | Methylation in Control | 7.36E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
TSS200 (cg24707219) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:4.26E-08; Z-score:1.60E+00 | ||
|
Methylation in Case |
8.74E-01 (Median) | Methylation in Control | 7.84E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg02867242) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:1.72E-28; Z-score:2.39E+00 | ||
|
Methylation in Case |
9.87E-01 (Median) | Methylation in Control | 9.22E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg10017105) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:5.49E-18; Z-score:3.82E+00 | ||
|
Methylation in Case |
8.99E-01 (Median) | Methylation in Control | 7.40E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg18949702) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:3.14E-13; Z-score:1.84E+00 | ||
|
Methylation in Case |
8.26E-01 (Median) | Methylation in Control | 7.74E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg14378924) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:4.85E-11; Z-score:1.98E+00 | ||
|
Methylation in Case |
7.37E-01 (Median) | Methylation in Control | 6.47E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg26472225) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.47E+00 | Statistic Test | p-value:1.18E-08; Z-score:3.18E+00 | ||
|
Methylation in Case |
4.45E-01 (Median) | Methylation in Control | 3.02E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg25368728) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:1.87E-08; Z-score:-2.38E+00 | ||
|
Methylation in Case |
7.84E-01 (Median) | Methylation in Control | 8.66E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg13691744) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.29E+00 | Statistic Test | p-value:2.23E-07; Z-score:1.51E+00 | ||
|
Methylation in Case |
2.37E-01 (Median) | Methylation in Control | 1.84E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg09153713) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:4.60E-07; Z-score:-2.21E+00 | ||
|
Methylation in Case |
6.26E-01 (Median) | Methylation in Control | 7.18E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg04489104) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.40E+00 | Statistic Test | p-value:5.38E-07; Z-score:2.11E+00 | ||
|
Methylation in Case |
2.14E-01 (Median) | Methylation in Control | 1.53E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg07546508) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:7.25E-07; Z-score:-1.67E+00 | ||
|
Methylation in Case |
7.18E-01 (Median) | Methylation in Control | 7.88E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg02775414) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:3.50E-06; Z-score:-1.26E+00 | ||
|
Methylation in Case |
7.39E-01 (Median) | Methylation in Control | 7.86E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg26138846) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:4.13E-06; Z-score:1.39E+00 | ||
|
Methylation in Case |
6.93E-01 (Median) | Methylation in Control | 6.26E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg15821546) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:4.15E-05; Z-score:-1.78E+00 | ||
|
Methylation in Case |
8.07E-01 (Median) | Methylation in Control | 8.65E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg27257955) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:6.92E-05; Z-score:-1.57E+00 | ||
|
Methylation in Case |
7.89E-01 (Median) | Methylation in Control | 8.41E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg04021562) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:2.52E-04; Z-score:1.02E+00 | ||
|
Methylation in Case |
3.86E-01 (Median) | Methylation in Control | 3.55E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg20961940) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:2.58E-04; Z-score:-1.58E+00 | ||
|
Methylation in Case |
7.30E-01 (Median) | Methylation in Control | 7.91E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg09298971) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:2.64E-04; Z-score:9.66E-01 | ||
|
Methylation in Case |
8.72E-01 (Median) | Methylation in Control | 8.58E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg18856043) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:2.65E-04; Z-score:9.07E-01 | ||
|
Methylation in Case |
8.04E-01 (Median) | Methylation in Control | 7.76E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg22882121) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:4.40E-04; Z-score:-1.39E+00 | ||
|
Methylation in Case |
8.60E-01 (Median) | Methylation in Control | 8.99E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg27005847) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:6.51E-04; Z-score:-1.64E+00 | ||
|
Methylation in Case |
8.22E-01 (Median) | Methylation in Control | 8.58E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg11726150) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:1.13E-03; Z-score:1.02E+00 | ||
|
Methylation in Case |
7.94E-01 (Median) | Methylation in Control | 7.64E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg05686323) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.19E-03; Z-score:-1.06E+00 | ||
|
Methylation in Case |
8.69E-01 (Median) | Methylation in Control | 8.91E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg19117051) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:1.32E-03; Z-score:7.17E-01 | ||
|
Methylation in Case |
8.80E-01 (Median) | Methylation in Control | 8.64E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg18263014) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:2.00E-03; Z-score:-9.76E-01 | ||
|
Methylation in Case |
8.92E-01 (Median) | Methylation in Control | 9.10E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg19117698) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:2.85E-03; Z-score:-8.95E-01 | ||
|
Methylation in Case |
8.35E-01 (Median) | Methylation in Control | 8.57E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg13975172) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:4.20E-03; Z-score:7.18E-01 | ||
|
Methylation in Case |
9.11E-01 (Median) | Methylation in Control | 8.95E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg20544437) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:4.54E-03; Z-score:-9.23E-01 | ||
|
Methylation in Case |
8.91E-01 (Median) | Methylation in Control | 9.16E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg27587935) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:7.34E-03; Z-score:-8.17E-01 | ||
|
Methylation in Case |
8.94E-01 (Median) | Methylation in Control | 9.15E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg24490633) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:1.30E-02; Z-score:-1.33E+00 | ||
|
Methylation in Case |
6.15E-01 (Median) | Methylation in Control | 6.71E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon37 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg26434400) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.79E-02; Z-score:-4.74E-01 | ||
|
Methylation in Case |
9.21E-01 (Median) | Methylation in Control | 9.29E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon38 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg07601645) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.37E-02; Z-score:-1.05E+00 | ||
|
Methylation in Case |
7.17E-01 (Median) | Methylation in Control | 7.53E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon39 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg01347702) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.39E-02; Z-score:-5.35E-01 | ||
|
Methylation in Case |
9.02E-01 (Median) | Methylation in Control | 9.12E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon40 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg25013052) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:3.59E-02; Z-score:-6.17E-01 | ||
|
Methylation in Case |
8.23E-01 (Median) | Methylation in Control | 8.36E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon41 |
Methylation of SLC44A4 in HIV infection | [ 5 ] | |||
|
Location |
Body (cg11640015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:3.97E-02; Z-score:4.28E-01 | ||
|
Methylation in Case |
8.70E-01 (Median) | Methylation in Control | 8.60E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
44 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
TSS1500 (cg16553272) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:4.54E-02; Z-score:6.11E-01 | ||
|
Methylation in Case |
8.63E-01 (Median) | Methylation in Control | 8.24E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
TSS200 (cg21236501) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:2.06E-05; Z-score:1.11E+00 | ||
|
Methylation in Case |
6.18E-01 (Median) | Methylation in Control | 5.61E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
TSS200 (cg04567302) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:2.06E-05; Z-score:1.03E+00 | ||
|
Methylation in Case |
7.52E-01 (Median) | Methylation in Control | 6.87E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
TSS200 (cg24529722) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:5.56E-05; Z-score:8.42E-01 | ||
|
Methylation in Case |
5.90E-01 (Median) | Methylation in Control | 5.39E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
TSS200 (cg03045620) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:1.86E-03; Z-score:9.41E-01 | ||
|
Methylation in Case |
5.87E-01 (Median) | Methylation in Control | 5.34E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
TSS200 (cg07363637) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:3.57E-02; Z-score:7.51E-01 | ||
|
Methylation in Case |
8.03E-01 (Median) | Methylation in Control | 7.67E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg07601645) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.51E+00 | Statistic Test | p-value:1.13E-24; Z-score:-3.61E+00 | ||
|
Methylation in Case |
4.20E-01 (Median) | Methylation in Control | 6.32E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg19955284) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.18E+00 | Statistic Test | p-value:3.52E-12; Z-score:2.09E+00 | ||
|
Methylation in Case |
8.09E-01 (Median) | Methylation in Control | 6.87E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg20961940) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.39E+00 | Statistic Test | p-value:8.48E-11; Z-score:-2.29E+00 | ||
|
Methylation in Case |
3.53E-01 (Median) | Methylation in Control | 4.89E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg07546508) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.30E+00 | Statistic Test | p-value:6.74E-10; Z-score:-1.82E+00 | ||
|
Methylation in Case |
4.02E-01 (Median) | Methylation in Control | 5.21E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg10969178) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.31E+00 | Statistic Test | p-value:1.05E-09; Z-score:-2.09E+00 | ||
|
Methylation in Case |
5.27E-01 (Median) | Methylation in Control | 6.89E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg27587935) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.38E+00 | Statistic Test | p-value:2.86E-08; Z-score:-1.49E+00 | ||
|
Methylation in Case |
3.79E-01 (Median) | Methylation in Control | 5.24E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg10951325) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:4.60E-07; Z-score:-1.81E+00 | ||
|
Methylation in Case |
5.72E-01 (Median) | Methylation in Control | 7.01E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg11640015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.29E+00 | Statistic Test | p-value:1.46E-06; Z-score:-1.68E+00 | ||
|
Methylation in Case |
4.75E-01 (Median) | Methylation in Control | 6.15E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg27005847) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:1.70E-06; Z-score:1.17E+00 | ||
|
Methylation in Case |
6.18E-01 (Median) | Methylation in Control | 5.69E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg25600103) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:1.80E-06; Z-score:-1.60E+00 | ||
|
Methylation in Case |
4.86E-01 (Median) | Methylation in Control | 5.75E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg11943056) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:4.72E-06; Z-score:1.27E+00 | ||
|
Methylation in Case |
7.34E-01 (Median) | Methylation in Control | 6.62E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg02775414) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:6.56E-06; Z-score:-1.48E+00 | ||
|
Methylation in Case |
4.07E-01 (Median) | Methylation in Control | 4.71E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg19853440) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:9.65E-06; Z-score:-1.42E+00 | ||
|
Methylation in Case |
3.75E-01 (Median) | Methylation in Control | 5.09E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg00031491) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.06E-05; Z-score:-9.61E-01 | ||
|
Methylation in Case |
8.27E-01 (Median) | Methylation in Control | 8.64E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg25368728) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.29E+00 | Statistic Test | p-value:1.18E-05; Z-score:-1.72E+00 | ||
|
Methylation in Case |
3.90E-01 (Median) | Methylation in Control | 5.02E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg07185041) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:2.18E-05; Z-score:1.52E+00 | ||
|
Methylation in Case |
7.86E-01 (Median) | Methylation in Control | 7.00E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg20498033) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:2.34E-05; Z-score:-1.32E+00 | ||
|
Methylation in Case |
8.56E-01 (Median) | Methylation in Control | 8.87E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg18263014) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:2.92E-05; Z-score:-1.02E+00 | ||
|
Methylation in Case |
8.91E-01 (Median) | Methylation in Control | 9.13E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg07705820) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.25E+00 | Statistic Test | p-value:9.70E-05; Z-score:-1.50E+00 | ||
|
Methylation in Case |
4.03E-01 (Median) | Methylation in Control | 5.04E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg11726150) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:1.98E-04; Z-score:1.10E+00 | ||
|
Methylation in Case |
7.44E-01 (Median) | Methylation in Control | 7.08E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg09153713) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:2.11E-04; Z-score:-7.25E-01 | ||
|
Methylation in Case |
3.63E-01 (Median) | Methylation in Control | 3.96E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg26434400) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:4.80E-04; Z-score:-1.18E+00 | ||
|
Methylation in Case |
4.95E-01 (Median) | Methylation in Control | 5.85E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg25013052) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:5.35E-04; Z-score:-7.59E-01 | ||
|
Methylation in Case |
3.44E-01 (Median) | Methylation in Control | 4.00E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg18445760) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:8.22E-04; Z-score:-8.57E-01 | ||
|
Methylation in Case |
8.50E-01 (Median) | Methylation in Control | 8.66E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg18856043) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:1.32E-03; Z-score:9.78E-01 | ||
|
Methylation in Case |
4.00E-01 (Median) | Methylation in Control | 3.51E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg01207036) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:1.79E-03; Z-score:-1.05E+00 | ||
|
Methylation in Case |
6.09E-01 (Median) | Methylation in Control | 6.58E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg04021562) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.23E+00 | Statistic Test | p-value:2.53E-03; Z-score:6.37E-01 | ||
|
Methylation in Case |
1.37E-01 (Median) | Methylation in Control | 1.11E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg07643404) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:4.40E-03; Z-score:6.88E-01 | ||
|
Methylation in Case |
8.62E-01 (Median) | Methylation in Control | 8.44E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg24732991) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:5.73E-03; Z-score:-4.56E-01 | ||
|
Methylation in Case |
8.68E-01 (Median) | Methylation in Control | 8.81E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg18949702) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:7.11E-03; Z-score:-5.79E-01 | ||
|
Methylation in Case |
8.44E-01 (Median) | Methylation in Control | 8.60E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon37 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg08506113) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:1.49E-02; Z-score:-8.15E-01 | ||
|
Methylation in Case |
3.79E-01 (Median) | Methylation in Control | 4.47E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon38 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg07438099) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:1.55E-02; Z-score:8.69E-01 | ||
|
Methylation in Case |
8.36E-01 (Median) | Methylation in Control | 8.08E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon39 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg00123181) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:1.65E-02; Z-score:1.08E+00 | ||
|
Methylation in Case |
8.83E-01 (Median) | Methylation in Control | 8.49E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon40 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg26138846) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:2.10E-02; Z-score:-5.24E-01 | ||
|
Methylation in Case |
7.54E-01 (Median) | Methylation in Control | 7.79E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon41 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg20544437) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:2.60E-02; Z-score:8.22E-01 | ||
|
Methylation in Case |
8.79E-01 (Median) | Methylation in Control | 8.44E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon42 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg17012525) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:2.99E-02; Z-score:-1.53E-01 | ||
|
Methylation in Case |
8.72E-01 (Median) | Methylation in Control | 8.75E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon43 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg11637059) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:3.59E-02; Z-score:-2.17E-03 | ||
|
Methylation in Case |
9.22E-01 (Median) | Methylation in Control | 9.22E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon44 |
Methylation of SLC44A4 in papillary thyroid cancer | [ 6 ] | |||
|
Location |
Body (cg05841219) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:4.45E-02; Z-score:6.84E-01 | ||
|
Methylation in Case |
7.25E-01 (Median) | Methylation in Control | 7.05E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
52 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
TSS200 (cg24707219) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:6.10E-06; Z-score:-1.33E+00 | ||
|
Methylation in Case |
7.10E-01 (Median) | Methylation in Control | 7.63E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
TSS200 (cg04567302) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:7.83E-06; Z-score:-8.07E-01 | ||
|
Methylation in Case |
6.95E-01 (Median) | Methylation in Control | 7.45E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
TSS200 (cg03045620) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:1.36E-05; Z-score:-1.31E+00 | ||
|
Methylation in Case |
5.12E-01 (Median) | Methylation in Control | 5.68E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
TSS200 (cg21236501) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:4.74E-05; Z-score:-1.28E+00 | ||
|
Methylation in Case |
5.52E-01 (Median) | Methylation in Control | 6.33E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
TSS200 (cg24529722) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:4.42E-03; Z-score:-6.89E-01 | ||
|
Methylation in Case |
5.23E-01 (Median) | Methylation in Control | 6.00E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
TSS200 (cg07363637) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:4.80E-03; Z-score:-5.91E-01 | ||
|
Methylation in Case |
6.20E-01 (Median) | Methylation in Control | 6.48E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg19955284) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:2.39E-20; Z-score:2.67E+00 | ||
|
Methylation in Case |
7.38E-01 (Median) | Methylation in Control | 6.19E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg00123181) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:3.67E-16; Z-score:2.07E+00 | ||
|
Methylation in Case |
8.10E-01 (Median) | Methylation in Control | 7.46E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg25600103) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:4.20E-16; Z-score:2.59E+00 | ||
|
Methylation in Case |
7.11E-01 (Median) | Methylation in Control | 5.97E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg14378924) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:6.54E-15; Z-score:2.15E+00 | ||
|
Methylation in Case |
5.43E-01 (Median) | Methylation in Control | 4.43E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg11640015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.23E+00 | Statistic Test | p-value:1.06E-14; Z-score:2.39E+00 | ||
|
Methylation in Case |
7.80E-01 (Median) | Methylation in Control | 6.35E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg04489104) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.51E+00 | Statistic Test | p-value:1.24E-12; Z-score:2.29E+00 | ||
|
Methylation in Case |
2.15E-01 (Median) | Methylation in Control | 1.42E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg24369466) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.17E+00 | Statistic Test | p-value:1.90E-12; Z-score:-4.02E+00 | ||
|
Methylation in Case |
7.60E-01 (Median) | Methylation in Control | 8.89E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg10017105) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.34E+00 | Statistic Test | p-value:2.10E-12; Z-score:1.62E+00 | ||
|
Methylation in Case |
7.63E-01 (Median) | Methylation in Control | 5.71E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg26472225) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.29E+00 | Statistic Test | p-value:1.25E-10; Z-score:2.67E+00 | ||
|
Methylation in Case |
3.88E-01 (Median) | Methylation in Control | 3.00E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg18949702) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:1.45E-10; Z-score:1.52E+00 | ||
|
Methylation in Case |
7.68E-01 (Median) | Methylation in Control | 7.16E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg00031491) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:1.46E-10; Z-score:1.32E+00 | ||
|
Methylation in Case |
8.08E-01 (Median) | Methylation in Control | 7.63E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg27005847) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.25E+00 | Statistic Test | p-value:1.74E-10; Z-score:-3.19E+00 | ||
|
Methylation in Case |
6.04E-01 (Median) | Methylation in Control | 7.57E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg07705820) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:4.71E-10; Z-score:-1.88E+00 | ||
|
Methylation in Case |
7.68E-01 (Median) | Methylation in Control | 8.13E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg22882121) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.28E+00 | Statistic Test | p-value:5.92E-10; Z-score:-3.37E+00 | ||
|
Methylation in Case |
5.55E-01 (Median) | Methylation in Control | 7.10E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg07438099) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:3.64E-09; Z-score:1.25E+00 | ||
|
Methylation in Case |
6.87E-01 (Median) | Methylation in Control | 6.23E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg07185041) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:1.11E-08; Z-score:-1.71E+00 | ||
|
Methylation in Case |
6.65E-01 (Median) | Methylation in Control | 7.47E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg26138846) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.20E+00 | Statistic Test | p-value:1.26E-08; Z-score:-2.11E+00 | ||
|
Methylation in Case |
5.59E-01 (Median) | Methylation in Control | 6.71E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg09130091) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:2.00E-08; Z-score:1.85E+00 | ||
|
Methylation in Case |
4.51E-01 (Median) | Methylation in Control | 4.07E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg19853440) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:9.57E-08; Z-score:1.98E+00 | ||
|
Methylation in Case |
6.99E-01 (Median) | Methylation in Control | 6.16E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg10969178) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.34E+00 | Statistic Test | p-value:1.27E-07; Z-score:2.70E+00 | ||
|
Methylation in Case |
8.88E-01 (Median) | Methylation in Control | 6.62E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg25013052) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:1.57E-07; Z-score:-1.29E+00 | ||
|
Methylation in Case |
7.48E-01 (Median) | Methylation in Control | 7.86E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg10951325) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:2.68E-06; Z-score:2.54E+00 | ||
|
Methylation in Case |
7.74E-01 (Median) | Methylation in Control | 6.59E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg11943056) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:4.59E-06; Z-score:-1.88E+00 | ||
|
Methylation in Case |
6.20E-01 (Median) | Methylation in Control | 7.12E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg05686323) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:1.94E-05; Z-score:-1.04E+00 | ||
|
Methylation in Case |
6.87E-01 (Median) | Methylation in Control | 7.36E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg09153713) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:2.02E-05; Z-score:-1.29E+00 | ||
|
Methylation in Case |
5.96E-01 (Median) | Methylation in Control | 6.55E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg07546508) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:2.63E-05; Z-score:-9.18E-01 | ||
|
Methylation in Case |
6.74E-01 (Median) | Methylation in Control | 7.14E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg18263014) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:5.06E-05; Z-score:-1.15E+00 | ||
|
Methylation in Case |
8.19E-01 (Median) | Methylation in Control | 8.60E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg11312554) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:5.43E-05; Z-score:8.88E-01 | ||
|
Methylation in Case |
8.48E-01 (Median) | Methylation in Control | 8.22E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg22872508) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:8.53E-05; Z-score:7.11E-01 | ||
|
Methylation in Case |
8.45E-01 (Median) | Methylation in Control | 8.21E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg02867242) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:1.93E-04; Z-score:3.43E+00 | ||
|
Methylation in Case |
9.31E-01 (Median) | Methylation in Control | 8.12E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon37 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg26434400) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:6.78E-04; Z-score:-4.92E-01 | ||
|
Methylation in Case |
8.86E-01 (Median) | Methylation in Control | 8.95E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon38 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg01347702) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:1.12E-03; Z-score:-1.30E+00 | ||
|
Methylation in Case |
5.73E-01 (Median) | Methylation in Control | 6.37E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon39 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg27587935) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.96E-03; Z-score:-6.52E-01 | ||
|
Methylation in Case |
8.37E-01 (Median) | Methylation in Control | 8.59E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon40 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg24732991) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:3.05E-03; Z-score:-7.19E-01 | ||
|
Methylation in Case |
8.33E-01 (Median) | Methylation in Control | 8.53E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon41 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg24490633) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:3.48E-03; Z-score:1.40E+00 | ||
|
Methylation in Case |
4.84E-01 (Median) | Methylation in Control | 4.35E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon42 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg20961940) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:4.12E-03; Z-score:-6.87E-01 | ||
|
Methylation in Case |
6.19E-01 (Median) | Methylation in Control | 6.52E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon43 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg09298971) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:6.02E-03; Z-score:-6.05E-01 | ||
|
Methylation in Case |
6.87E-01 (Median) | Methylation in Control | 7.49E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon44 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg19117698) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:7.03E-03; Z-score:2.20E-01 | ||
|
Methylation in Case |
7.20E-01 (Median) | Methylation in Control | 6.94E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon45 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg13691744) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.47E+00 | Statistic Test | p-value:7.92E-03; Z-score:-1.22E+00 | ||
|
Methylation in Case |
2.49E-01 (Median) | Methylation in Control | 3.66E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon46 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg23701033) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:8.84E-03; Z-score:-4.62E-01 | ||
|
Methylation in Case |
5.99E-01 (Median) | Methylation in Control | 6.25E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon47 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg15821546) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:9.12E-03; Z-score:-1.06E+00 | ||
|
Methylation in Case |
5.33E-01 (Median) | Methylation in Control | 5.74E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon48 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg27257955) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:1.45E-02; Z-score:5.25E-01 | ||
|
Methylation in Case |
6.53E-01 (Median) | Methylation in Control | 6.08E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon49 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg02775414) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:2.12E-02; Z-score:3.96E-01 | ||
|
Methylation in Case |
6.18E-01 (Median) | Methylation in Control | 6.02E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon50 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg04261164) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:2.26E-02; Z-score:5.75E-01 | ||
|
Methylation in Case |
8.44E-01 (Median) | Methylation in Control | 8.13E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon51 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg02721751) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:4.55E-02; Z-score:9.11E-01 | ||
|
Methylation in Case |
8.17E-01 (Median) | Methylation in Control | 7.78E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon52 |
Methylation of SLC44A4 in breast cancer | [ 7 ] | |||
|
Location |
Body (cg13975172) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:4.71E-02; Z-score:6.78E-01 | ||
|
Methylation in Case |
7.78E-01 (Median) | Methylation in Control | 7.48E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
50 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg00964484) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.50E+00 | Statistic Test | p-value:2.37E-14; Z-score:2.15E+00 | ||
|
Methylation in Case |
5.51E-01 (Median) | Methylation in Control | 3.66E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg13280283) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+01 | Statistic Test | p-value:4.51E-14; Z-score:1.56E+01 | ||
|
Methylation in Case |
3.24E-01 (Median) | Methylation in Control | 3.20E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg07363637) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:1.89E-06; Z-score:1.04E+00 | ||
|
Methylation in Case |
7.69E-01 (Median) | Methylation in Control | 7.35E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg21236501) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:5.97E-06; Z-score:1.60E+00 | ||
|
Methylation in Case |
7.33E-01 (Median) | Methylation in Control | 6.89E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg24707219) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:1.75E-03; Z-score:8.48E-01 | ||
|
Methylation in Case |
8.47E-01 (Median) | Methylation in Control | 8.25E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg24529722) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:3.44E-03; Z-score:1.03E+00 | ||
|
Methylation in Case |
7.03E-01 (Median) | Methylation in Control | 6.69E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg04567302) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:2.70E-02; Z-score:7.28E-01 | ||
|
Methylation in Case |
8.30E-01 (Median) | Methylation in Control | 8.06E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
1stExon (cg06123346) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.52E+00 | Statistic Test | p-value:5.18E-16; Z-score:-3.88E+00 | ||
|
Methylation in Case |
3.02E-01 (Median) | Methylation in Control | 4.60E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
1stExon (cg13223402) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:5.57E+00 | Statistic Test | p-value:2.60E-13; Z-score:8.19E+00 | ||
|
Methylation in Case |
2.24E-01 (Median) | Methylation in Control | 4.02E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg01454305) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.51E+00 | Statistic Test | p-value:1.46E-18; Z-score:-4.27E+00 | ||
|
Methylation in Case |
4.46E-01 (Median) | Methylation in Control | 6.72E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg14495732) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.71E+00 | Statistic Test | p-value:1.73E-17; Z-score:-6.67E+00 | ||
|
Methylation in Case |
3.94E-01 (Median) | Methylation in Control | 6.74E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg18426477) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.31E+00 | Statistic Test | p-value:1.59E-15; Z-score:-3.22E+00 | ||
|
Methylation in Case |
4.65E-01 (Median) | Methylation in Control | 6.07E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg23930334) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-5.72E+00 | Statistic Test | p-value:1.93E-15; Z-score:-2.58E+00 | ||
|
Methylation in Case |
8.45E-02 (Median) | Methylation in Control | 4.83E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg22891413) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.34E+00 | Statistic Test | p-value:4.30E-15; Z-score:-5.10E+00 | ||
|
Methylation in Case |
6.68E-01 (Median) | Methylation in Control | 8.92E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg02483462) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:1.69E-12; Z-score:-5.04E+00 | ||
|
Methylation in Case |
4.60E-01 (Median) | Methylation in Control | 6.25E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg22792910) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.49E+00 | Statistic Test | p-value:1.63E-11; Z-score:-1.80E+00 | ||
|
Methylation in Case |
2.89E-01 (Median) | Methylation in Control | 4.32E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg07546508) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.37E+00 | Statistic Test | p-value:2.59E-09; Z-score:-1.90E+00 | ||
|
Methylation in Case |
3.37E-01 (Median) | Methylation in Control | 4.62E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg15821546) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:3.63E-08; Z-score:-1.33E+00 | ||
|
Methylation in Case |
6.08E-01 (Median) | Methylation in Control | 6.79E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg19853440) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:6.60E-07; Z-score:-1.06E+00 | ||
|
Methylation in Case |
7.15E-01 (Median) | Methylation in Control | 7.42E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg14378924) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:7.90E-07; Z-score:-1.35E+00 | ||
|
Methylation in Case |
5.23E-01 (Median) | Methylation in Control | 5.90E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg00123181) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:3.00E-06; Z-score:-7.68E-01 | ||
|
Methylation in Case |
7.82E-01 (Median) | Methylation in Control | 8.06E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg21399428) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:3.71E-06; Z-score:-1.45E+00 | ||
|
Methylation in Case |
7.94E-01 (Median) | Methylation in Control | 8.19E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg24732991) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.30E+00 | Statistic Test | p-value:3.93E-06; Z-score:-1.53E+00 | ||
|
Methylation in Case |
4.58E-01 (Median) | Methylation in Control | 5.94E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg02252769) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:1.67E-05; Z-score:-1.23E+00 | ||
|
Methylation in Case |
7.10E-01 (Median) | Methylation in Control | 7.46E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg18856043) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:2.55E-05; Z-score:-1.18E+00 | ||
|
Methylation in Case |
4.88E-01 (Median) | Methylation in Control | 5.61E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg11312554) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:5.19E-05; Z-score:-8.77E-01 | ||
|
Methylation in Case |
8.53E-01 (Median) | Methylation in Control | 8.72E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg11637059) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:8.27E-05; Z-score:-7.10E-01 | ||
|
Methylation in Case |
8.25E-01 (Median) | Methylation in Control | 8.42E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg18445760) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.47E-04; Z-score:-4.24E-01 | ||
|
Methylation in Case |
8.65E-01 (Median) | Methylation in Control | 8.75E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg09130091) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:4.10E-04; Z-score:-9.26E-01 | ||
|
Methylation in Case |
4.52E-01 (Median) | Methylation in Control | 4.88E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg07185041) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:8.05E-04; Z-score:-6.51E-01 | ||
|
Methylation in Case |
6.96E-01 (Median) | Methylation in Control | 7.42E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg13975172) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.99E-03; Z-score:-3.23E-01 | ||
|
Methylation in Case |
8.23E-01 (Median) | Methylation in Control | 8.34E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg05841219) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.01E-03; Z-score:-1.28E-01 | ||
|
Methylation in Case |
8.57E-01 (Median) | Methylation in Control | 8.62E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg17012525) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.47E-03; Z-score:-2.87E-01 | ||
|
Methylation in Case |
8.20E-01 (Median) | Methylation in Control | 8.27E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg02867242) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.98E-03; Z-score:-4.49E-01 | ||
|
Methylation in Case |
9.61E-01 (Median) | Methylation in Control | 9.67E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg13691744) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:3.85E-03; Z-score:-9.36E-01 | ||
|
Methylation in Case |
3.57E-01 (Median) | Methylation in Control | 4.07E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg22882121) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:5.16E-03; Z-score:-1.80E-01 | ||
|
Methylation in Case |
7.79E-01 (Median) | Methylation in Control | 7.87E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon37 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg21618695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:7.40E-03; Z-score:-8.06E-01 | ||
|
Methylation in Case |
8.06E-01 (Median) | Methylation in Control | 8.18E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon38 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg01347702) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:7.88E-03; Z-score:-7.98E-01 | ||
|
Methylation in Case |
6.16E-01 (Median) | Methylation in Control | 6.78E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon39 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg19117698) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:8.43E-03; Z-score:6.25E-01 | ||
|
Methylation in Case |
7.67E-01 (Median) | Methylation in Control | 7.47E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon40 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg25013052) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.31E-02; Z-score:-3.01E-01 | ||
|
Methylation in Case |
7.36E-01 (Median) | Methylation in Control | 7.48E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon41 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg10017105) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.48E-02; Z-score:-3.16E-01 | ||
|
Methylation in Case |
8.85E-01 (Median) | Methylation in Control | 8.97E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon42 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg19117051) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:1.53E-02; Z-score:3.42E-01 | ||
|
Methylation in Case |
7.15E-01 (Median) | Methylation in Control | 6.84E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon43 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg02721751) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.61E-02; Z-score:-3.84E-01 | ||
|
Methylation in Case |
8.38E-01 (Median) | Methylation in Control | 8.58E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon44 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg08506113) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.94E-02; Z-score:-2.73E-01 | ||
|
Methylation in Case |
8.48E-01 (Median) | Methylation in Control | 8.55E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon45 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg18949702) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.97E-02; Z-score:-2.83E-01 | ||
|
Methylation in Case |
7.89E-01 (Median) | Methylation in Control | 7.94E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon46 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg00031491) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:2.22E-02; Z-score:-1.26E-01 | ||
|
Methylation in Case |
8.01E-01 (Median) | Methylation in Control | 8.14E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon47 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg09298971) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:3.33E-02; Z-score:3.58E-01 | ||
|
Methylation in Case |
8.36E-01 (Median) | Methylation in Control | 8.28E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon48 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg27257955) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:4.29E-02; Z-score:-4.79E-01 | ||
|
Methylation in Case |
6.55E-01 (Median) | Methylation in Control | 6.75E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon49 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg04489104) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:4.83E-02; Z-score:-8.74E-01 | ||
|
Methylation in Case |
2.56E-01 (Median) | Methylation in Control | 2.98E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon50 |
Methylation of SLC44A4 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg04261164) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:4.87E-02; Z-score:-1.78E-01 | ||
|
Methylation in Case |
8.70E-01 (Median) | Methylation in Control | 8.73E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
10 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC44A4 in panic disorder | [ 9 ] | |||
|
Location |
TSS200 (cg04567302) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.29E+00 | Statistic Test | p-value:3.10E-02; Z-score:-3.81E-01 | ||
|
Methylation in Case |
7.54E-01 (Median) | Methylation in Control | 9.69E-01 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC44A4 in panic disorder | [ 9 ] | |||
|
Location |
Body (cg02775414) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:5.48E-04; Z-score:6.87E-01 | ||
|
Methylation in Case |
2.03E+00 (Median) | Methylation in Control | 1.71E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC44A4 in panic disorder | [ 9 ] | |||
|
Location |
Body (cg26472225) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-8.77E-01 | Statistic Test | p-value:6.79E-04; Z-score:-6.51E-01 | ||
|
Methylation in Case |
-1.84E+00 (Median) | Methylation in Control | -1.61E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC44A4 in panic disorder | [ 9 ] | |||
|
Location |
Body (cg07205860) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:6.90E-03; Z-score:3.88E-01 | ||
|
Methylation in Case |
3.01E+00 (Median) | Methylation in Control | 2.92E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC44A4 in panic disorder | [ 9 ] | |||
|
Location |
Body (cg11312554) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:1.05E-02; Z-score:1.81E-01 | ||
|
Methylation in Case |
3.54E+00 (Median) | Methylation in Control | 3.49E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC44A4 in panic disorder | [ 9 ] | |||
|
Location |
Body (cg02867242) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:1.51E-02; Z-score:-3.39E-01 | ||
|
Methylation in Case |
3.82E+00 (Median) | Methylation in Control | 4.08E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC44A4 in panic disorder | [ 9 ] | |||
|
Location |
Body (cg24490633) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.28E+00 | Statistic Test | p-value:2.41E-02; Z-score:4.54E-01 | ||
|
Methylation in Case |
7.80E-01 (Median) | Methylation in Control | 6.09E-01 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC44A4 in panic disorder | [ 9 ] | |||
|
Location |
Body (cg11637059) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:4.08E-02; Z-score:-5.29E-01 | ||
|
Methylation in Case |
3.39E+00 (Median) | Methylation in Control | 3.54E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC44A4 in panic disorder | [ 9 ] | |||
|
Location |
Body (cg07643404) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:4.74E-02; Z-score:2.18E-01 | ||
|
Methylation in Case |
3.18E+00 (Median) | Methylation in Control | 3.10E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC44A4 in panic disorder | [ 9 ] | |||
|
Location |
Body (cg10017105) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:4.89E-02; Z-score:-3.86E-01 | ||
|
Methylation in Case |
1.58E+00 (Median) | Methylation in Control | 1.74E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Atypical teratoid rhabdoid tumor |
35 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg00031491) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:1.50E-05; Z-score:-1.08E+00 | ||
|
Methylation in Case |
7.62E-01 (Median) | Methylation in Control | 8.54E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg00123181) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:1.71E-05; Z-score:-1.09E+00 | ||
|
Methylation in Case |
7.61E-01 (Median) | Methylation in Control | 8.26E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg01207036) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.24E+00 | Statistic Test | p-value:5.59E-05; Z-score:6.85E-01 | ||
|
Methylation in Case |
6.31E-01 (Median) | Methylation in Control | 5.08E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg01347702) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:6.17E-05; Z-score:-1.14E+00 | ||
|
Methylation in Case |
5.47E-01 (Median) | Methylation in Control | 6.32E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg02252769) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.27E+00 | Statistic Test | p-value:1.36E-04; Z-score:-6.49E-01 | ||
|
Methylation in Case |
3.89E-01 (Median) | Methylation in Control | 4.96E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg02721751) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:2.23E-04; Z-score:-8.87E-01 | ||
|
Methylation in Case |
8.58E-01 (Median) | Methylation in Control | 8.91E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg02775414) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:2.28E-04; Z-score:8.89E-01 | ||
|
Methylation in Case |
8.87E-01 (Median) | Methylation in Control | 7.44E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg02867242) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:2.31E-04; Z-score:-8.97E-01 | ||
|
Methylation in Case |
7.93E-01 (Median) | Methylation in Control | 8.63E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg04021562) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.20E+00 | Statistic Test | p-value:5.28E-04; Z-score:-7.74E-01 | ||
|
Methylation in Case |
4.59E-01 (Median) | Methylation in Control | 5.53E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg04261164) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.62E+00 | Statistic Test | p-value:6.48E-04; Z-score:-9.70E-01 | ||
|
Methylation in Case |
1.89E-01 (Median) | Methylation in Control | 3.05E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg04489104) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:8.27E-04; Z-score:9.38E-01 | ||
|
Methylation in Case |
6.83E-01 (Median) | Methylation in Control | 6.17E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg05686323) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.63E-03; Z-score:-5.85E-01 | ||
|
Methylation in Case |
8.52E-01 (Median) | Methylation in Control | 8.88E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg05841219) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:1.72E-03; Z-score:-4.41E-01 | ||
|
Methylation in Case |
1.13E-01 (Median) | Methylation in Control | 1.26E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg07185041) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:3.42E-03; Z-score:-4.67E-01 | ||
|
Methylation in Case |
7.48E-01 (Median) | Methylation in Control | 7.61E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg07205860) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.16E+00 | Statistic Test | p-value:3.55E-03; Z-score:6.11E-01 | ||
|
Methylation in Case |
3.01E-01 (Median) | Methylation in Control | 2.58E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg07438099) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.60E+00 | Statistic Test | p-value:3.91E-03; Z-score:-3.21E-01 | ||
|
Methylation in Case |
6.71E-02 (Median) | Methylation in Control | 1.08E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg07546508) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:4.27E-03; Z-score:5.49E-01 | ||
|
Methylation in Case |
8.06E-01 (Median) | Methylation in Control | 7.67E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg07601645) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:4.49E-03; Z-score:-6.87E-01 | ||
|
Methylation in Case |
6.92E-01 (Median) | Methylation in Control | 7.90E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg07643404) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.67E+00 | Statistic Test | p-value:4.67E-03; Z-score:-5.25E-01 | ||
|
Methylation in Case |
1.24E-01 (Median) | Methylation in Control | 2.07E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg07705820) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:4.91E-03; Z-score:-4.04E-01 | ||
|
Methylation in Case |
1.51E-01 (Median) | Methylation in Control | 1.78E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg08506113) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:7.19E-03; Z-score:-3.91E-01 | ||
|
Methylation in Case |
8.15E-01 (Median) | Methylation in Control | 8.38E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg09130091) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.51E+00 | Statistic Test | p-value:9.29E-03; Z-score:-3.46E-01 | ||
|
Methylation in Case |
6.84E-02 (Median) | Methylation in Control | 1.03E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg09153713) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:9.43E-03; Z-score:5.02E-01 | ||
|
Methylation in Case |
9.06E-01 (Median) | Methylation in Control | 8.81E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg09298971) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:9.79E-03; Z-score:-4.46E-01 | ||
|
Methylation in Case |
1.62E-01 (Median) | Methylation in Control | 1.87E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg10017105) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:1.34E-02; Z-score:1.70E-01 | ||
|
Methylation in Case |
5.39E-02 (Median) | Methylation in Control | 4.86E-02 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg10951325) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:1.93E-02; Z-score:5.56E-01 | ||
|
Methylation in Case |
8.97E-01 (Median) | Methylation in Control | 8.67E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg10969178) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:1.95E-02; Z-score:6.56E-01 | ||
|
Methylation in Case |
8.62E-01 (Median) | Methylation in Control | 7.70E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg11312554) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:2.12E-02; Z-score:-3.53E-01 | ||
|
Methylation in Case |
7.81E-01 (Median) | Methylation in Control | 8.13E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg11637059) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:2.39E-02; Z-score:-5.74E-01 | ||
|
Methylation in Case |
7.73E-01 (Median) | Methylation in Control | 8.45E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg11640015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:2.40E-02; Z-score:6.36E-01 | ||
|
Methylation in Case |
9.12E-01 (Median) | Methylation in Control | 8.72E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg11726150) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:2.49E-02; Z-score:-5.82E-01 | ||
|
Methylation in Case |
8.65E-01 (Median) | Methylation in Control | 8.87E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg11943056) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:2.59E-02; Z-score:7.39E-01 | ||
|
Methylation in Case |
8.36E-01 (Median) | Methylation in Control | 7.53E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg13691744) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:3.82E-02; Z-score:4.98E-01 | ||
|
Methylation in Case |
8.73E-01 (Median) | Methylation in Control | 8.28E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg13975172) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:4.08E-02; Z-score:-3.94E-01 | ||
|
Methylation in Case |
8.56E-01 (Median) | Methylation in Control | 8.81E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC44A4 in atypical teratoid rhabdoid tumor | [ 10 ] | |||
|
Location |
Body (cg14378924) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:4.60E-02; Z-score:2.11E-01 | ||
|
Methylation in Case |
8.36E-01 (Median) | Methylation in Control | 8.22E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Celiac disease |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC44A4 in celiac disease | [ 11 ] | |||
|
Location |
Body (cg11640015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.89E-02; Z-score:-4.92E-01 | ||
|
Methylation in Case |
8.72E-01 (Median) | Methylation in Control | 8.95E-01 (Median) | ||
|
Studied Phenotype |
Celiac disease[ ICD-11:DA95] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC44A4 in clear cell renal cell carcinoma | [ 12 ] | |||
|
Location |
Body (cg20961940) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.23E+00 | Statistic Test | p-value:2.32E-04; Z-score:-1.94E+00 | ||
|
Methylation in Case |
4.85E-01 (Median) | Methylation in Control | 5.98E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC44A4 in clear cell renal cell carcinoma | [ 12 ] | |||
|
Location |
Body (cg21399428) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:1.34E-02; Z-score:7.54E-01 | ||
|
Methylation in Case |
8.96E-01 (Median) | Methylation in Control | 8.78E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
28 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg04489104) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.41E+00 | Statistic Test | p-value:7.47E-08; Z-score:-1.72E+00 | ||
|
Methylation in Case |
3.63E-01 (Median) | Methylation in Control | 5.11E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg20961940) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:1.17E-05; Z-score:-1.42E+00 | ||
|
Methylation in Case |
6.88E-01 (Median) | Methylation in Control | 7.72E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg02867242) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.93E-05; Z-score:-1.66E+00 | ||
|
Methylation in Case |
9.70E-01 (Median) | Methylation in Control | 9.81E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg05686323) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:2.26E-04; Z-score:-9.73E-01 | ||
|
Methylation in Case |
7.48E-01 (Median) | Methylation in Control | 8.10E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg23701033) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:2.31E-04; Z-score:-8.20E-01 | ||
|
Methylation in Case |
7.24E-01 (Median) | Methylation in Control | 7.67E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg20498033) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:6.99E-04; Z-score:-5.75E-01 | ||
|
Methylation in Case |
9.03E-01 (Median) | Methylation in Control | 9.32E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg18949702) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.28E-03; Z-score:-3.91E-01 | ||
|
Methylation in Case |
9.20E-01 (Median) | Methylation in Control | 9.25E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg22872508) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.73E-03; Z-score:-7.26E-01 | ||
|
Methylation in Case |
9.22E-01 (Median) | Methylation in Control | 9.33E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg02775414) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:4.74E-03; Z-score:-8.19E-01 | ||
|
Methylation in Case |
7.40E-01 (Median) | Methylation in Control | 7.90E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg19117698) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:4.99E-03; Z-score:9.01E-01 | ||
|
Methylation in Case |
8.84E-01 (Median) | Methylation in Control | 8.66E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg10017105) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:5.70E-03; Z-score:-8.16E-01 | ||
|
Methylation in Case |
9.01E-01 (Median) | Methylation in Control | 9.34E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg09153713) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:5.80E-03; Z-score:-7.40E-01 | ||
|
Methylation in Case |
7.35E-01 (Median) | Methylation in Control | 7.74E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg11640015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:5.91E-03; Z-score:-2.88E-01 | ||
|
Methylation in Case |
9.33E-01 (Median) | Methylation in Control | 9.36E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg07705820) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:7.81E-03; Z-score:-5.47E-01 | ||
|
Methylation in Case |
8.91E-01 (Median) | Methylation in Control | 9.16E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg07546508) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:8.44E-03; Z-score:-6.25E-01 | ||
|
Methylation in Case |
7.03E-01 (Median) | Methylation in Control | 7.43E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg15821546) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.25E+00 | Statistic Test | p-value:1.16E-02; Z-score:1.03E+00 | ||
|
Methylation in Case |
6.15E-01 (Median) | Methylation in Control | 4.92E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg26138846) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:1.25E-02; Z-score:1.11E+00 | ||
|
Methylation in Case |
7.01E-01 (Median) | Methylation in Control | 5.73E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg25368728) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.38E-02; Z-score:-5.22E-01 | ||
|
Methylation in Case |
8.51E-01 (Median) | Methylation in Control | 8.83E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg22882121) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:1.42E-02; Z-score:1.15E+00 | ||
|
Methylation in Case |
7.39E-01 (Median) | Methylation in Control | 6.08E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg09298971) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.18E+00 | Statistic Test | p-value:1.68E-02; Z-score:4.44E-01 | ||
|
Methylation in Case |
4.89E-01 (Median) | Methylation in Control | 4.14E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg26472225) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:1.99E-02; Z-score:-5.37E-01 | ||
|
Methylation in Case |
7.06E-01 (Median) | Methylation in Control | 7.54E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg24732991) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.99E-02; Z-score:-4.02E-01 | ||
|
Methylation in Case |
8.98E-01 (Median) | Methylation in Control | 9.19E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg18263014) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:2.41E-02; Z-score:-3.14E-01 | ||
|
Methylation in Case |
9.06E-01 (Median) | Methylation in Control | 9.21E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg24369466) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:2.80E-02; Z-score:9.18E-01 | ||
|
Methylation in Case |
7.33E-01 (Median) | Methylation in Control | 6.27E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg10969178) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:3.45E-02; Z-score:-2.09E-01 | ||
|
Methylation in Case |
9.58E-01 (Median) | Methylation in Control | 9.60E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg21399428) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:3.47E-02; Z-score:-2.51E-01 | ||
|
Methylation in Case |
9.22E-01 (Median) | Methylation in Control | 9.25E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC44A4 in colorectal cancer | [ 13 ] | |||
|
Location |
Body (cg07438099) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.53E-02; Z-score:-2.72E-01 | ||
|
Methylation in Case |
7.94E-01 (Median) | Methylation in Control | 8.03E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Depression |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC44A4 in depression | [ 14 ] | |||
|
Location |
Body (cg22882121) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:7.77E-03; Z-score:-3.12E-01 | ||
|
Methylation in Case |
8.36E-01 (Median) | Methylation in Control | 8.44E-01 (Median) | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC44A4 in depression | [ 14 ] | |||
|
Location |
Body (cg04261164) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.91E-02; Z-score:-4.98E-01 | ||
|
Methylation in Case |
8.62E-01 (Median) | Methylation in Control | 8.70E-01 (Median) | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
14 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC44A4 in lung adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg07546508) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.68E-04; Z-score:-1.95E+00 | ||
|
Methylation in Case |
7.39E-01 (Median) | Methylation in Control | 7.83E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC44A4 in lung adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg20961940) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:7.92E-04; Z-score:-2.34E+00 | ||
|
Methylation in Case |
6.79E-01 (Median) | Methylation in Control | 7.44E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC44A4 in lung adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg07705820) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:8.31E-04; Z-score:-1.70E+00 | ||
|
Methylation in Case |
8.28E-01 (Median) | Methylation in Control | 8.54E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC44A4 in lung adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg25368728) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:3.51E-03; Z-score:-1.55E+00 | ||
|
Methylation in Case |
8.00E-01 (Median) | Methylation in Control | 8.31E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC44A4 in lung adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg19955284) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:5.21E-03; Z-score:2.23E+00 | ||
|
Methylation in Case |
8.55E-01 (Median) | Methylation in Control | 7.94E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC44A4 in lung adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg19117051) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:7.73E-03; Z-score:1.85E+00 | ||
|
Methylation in Case |
7.50E-01 (Median) | Methylation in Control | 6.43E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC44A4 in lung adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg08506113) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:9.01E-03; Z-score:-1.51E+00 | ||
|
Methylation in Case |
8.57E-01 (Median) | Methylation in Control | 8.86E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC44A4 in lung adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg11726150) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.25E+00 | Statistic Test | p-value:9.34E-03; Z-score:1.92E+00 | ||
|
Methylation in Case |
6.46E-01 (Median) | Methylation in Control | 5.16E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC44A4 in lung adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg18263014) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.05E-02; Z-score:-2.10E+00 | ||
|
Methylation in Case |
8.72E-01 (Median) | Methylation in Control | 8.92E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC44A4 in lung adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg05686323) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.30E-02; Z-score:-1.77E+00 | ||
|
Methylation in Case |
7.79E-01 (Median) | Methylation in Control | 8.26E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC44A4 in lung adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg09153713) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:1.44E-02; Z-score:-1.12E+00 | ||
|
Methylation in Case |
6.84E-01 (Median) | Methylation in Control | 7.19E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC44A4 in lung adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg04261164) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:1.83E-02; Z-score:1.83E+00 | ||
|
Methylation in Case |
8.96E-01 (Median) | Methylation in Control | 8.55E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC44A4 in lung adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg25013052) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:2.30E-02; Z-score:-1.09E+00 | ||
|
Methylation in Case |
7.96E-01 (Median) | Methylation in Control | 8.20E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC44A4 in lung adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg14378924) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:3.03E-02; Z-score:1.17E+00 | ||
|
Methylation in Case |
6.42E-01 (Median) | Methylation in Control | 5.99E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Systemic lupus erythematosus |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC44A4 in systemic lupus erythematosus | [ 16 ] | |||
|
Location |
Body (cg05841219) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:3.28E-03; Z-score:-3.09E-01 | ||
|
Methylation in Case |
8.02E-01 (Median) | Methylation in Control | 8.15E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC44A4 in systemic lupus erythematosus | [ 16 ] | |||
|
Location |
Body (cg20544437) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.59E-02; Z-score:-1.65E-01 | ||
|
Methylation in Case |
8.70E-01 (Median) | Methylation in Control | 8.76E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC44A4 in systemic lupus erythematosus | [ 16 ] | |||
|
Location |
Body (cg18263014) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.96E-02; Z-score:-1.82E-01 | ||
|
Methylation in Case |
8.77E-01 (Median) | Methylation in Control | 8.85E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC44A4 in systemic lupus erythematosus | [ 16 ] | |||
|
Location |
Body (cg11637059) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.24E-02; Z-score:-1.83E-01 | ||
|
Methylation in Case |
9.11E-01 (Median) | Methylation in Control | 9.16E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC44A4 in systemic lupus erythematosus | [ 16 ] | |||
|
Location |
Body (cg10969178) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:4.73E-02; Z-score:-6.54E-02 | ||
|
Methylation in Case |
9.53E-01 (Median) | Methylation in Control | 9.54E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Multilayered rosettes embryonal tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypermethylation of SLC44A4 in multilayered rosettes embryonal tumour than that in healthy individual | ||||
Studied Phenotype |
Multilayered rosettes embryonal tumour [ICD-11:2A00.1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.002165926; Fold-change:0.207396915; Z-score:0.977940417 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
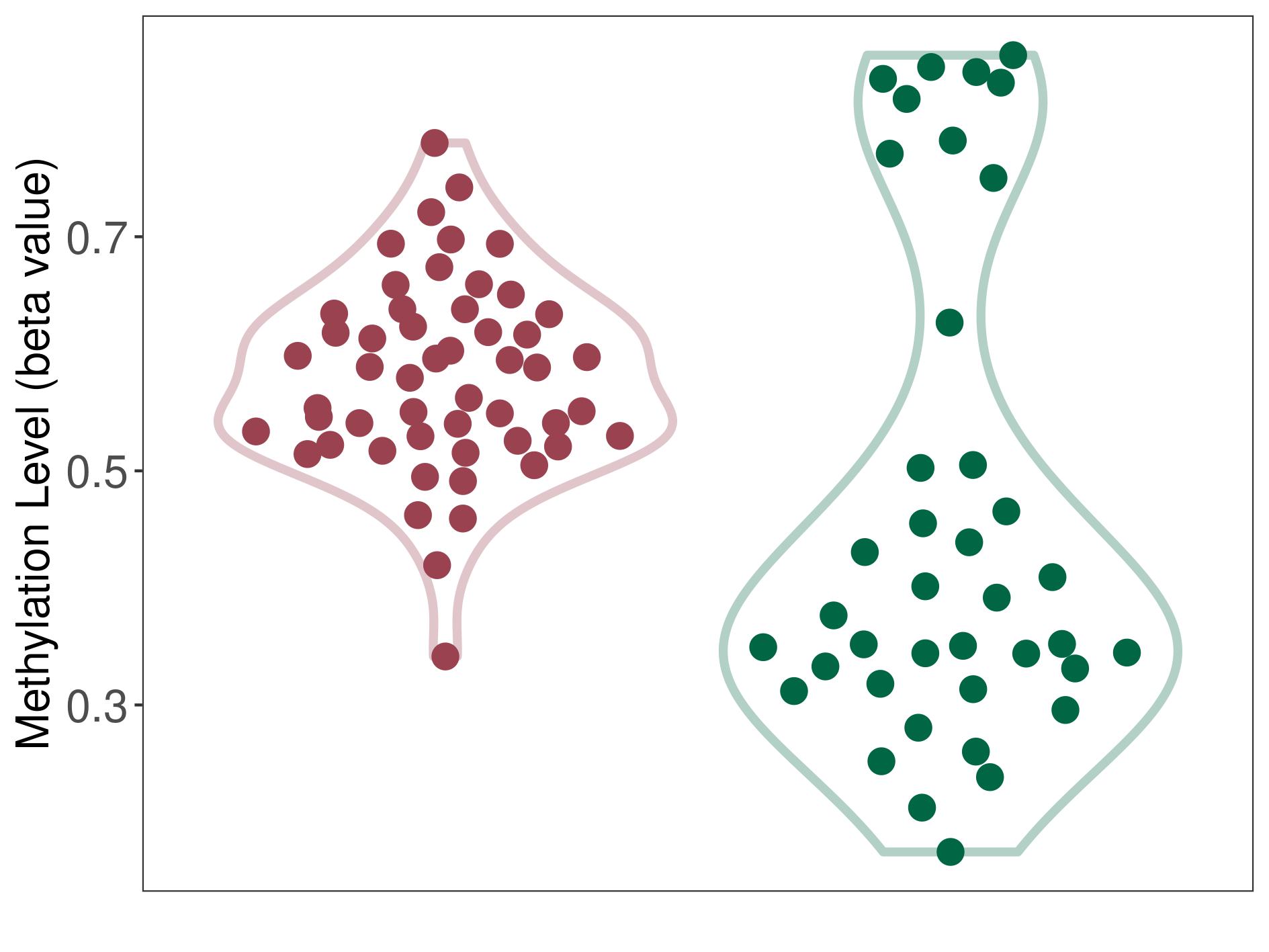
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Oligodendroglioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypermethylation of SLC44A4 in oligodendroglioma than that in healthy individual | ||||
Studied Phenotype |
Oligodendroglioma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.44E-07; Fold-change:0.224388524; Z-score:1.066235898 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
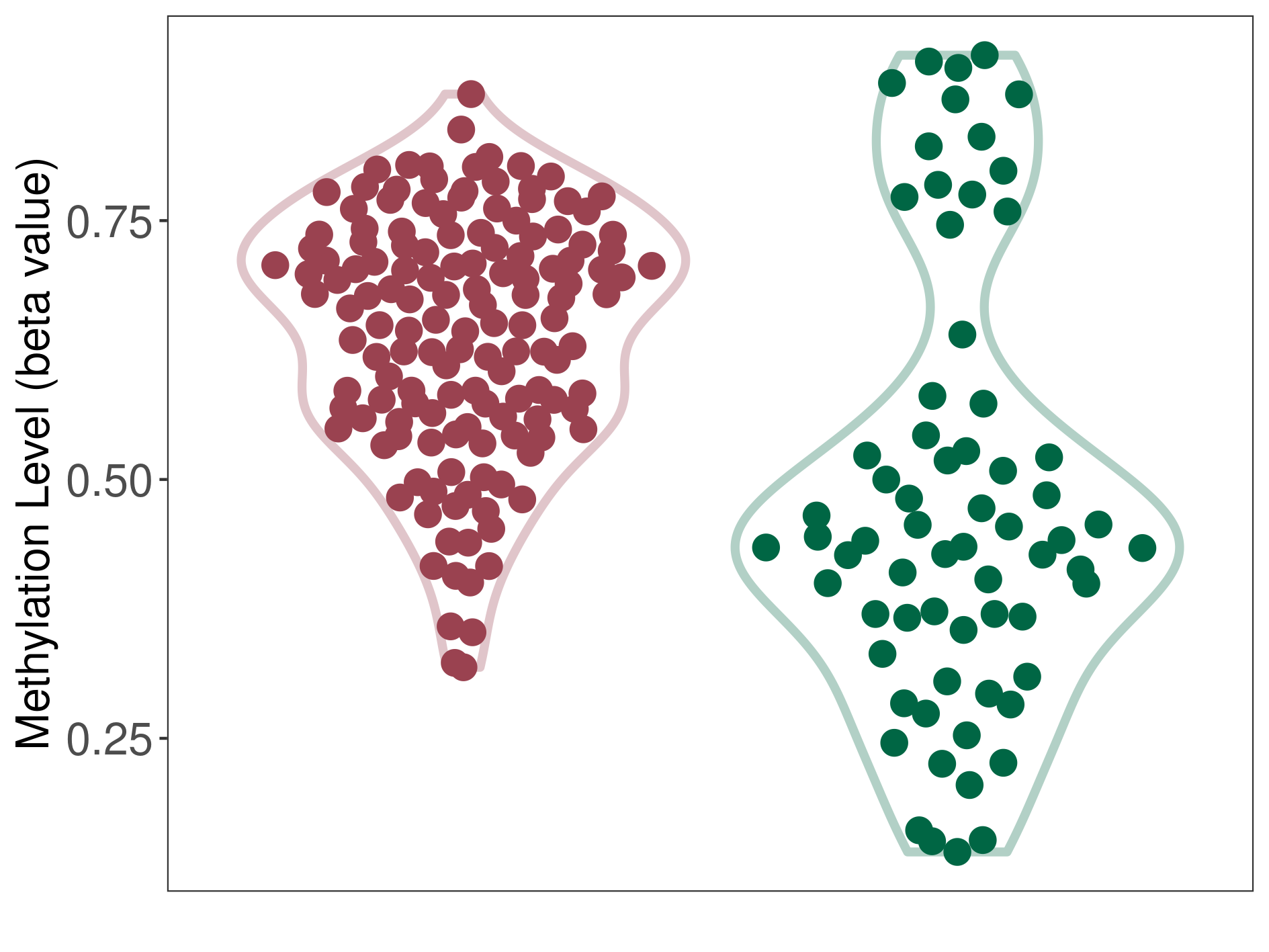
|
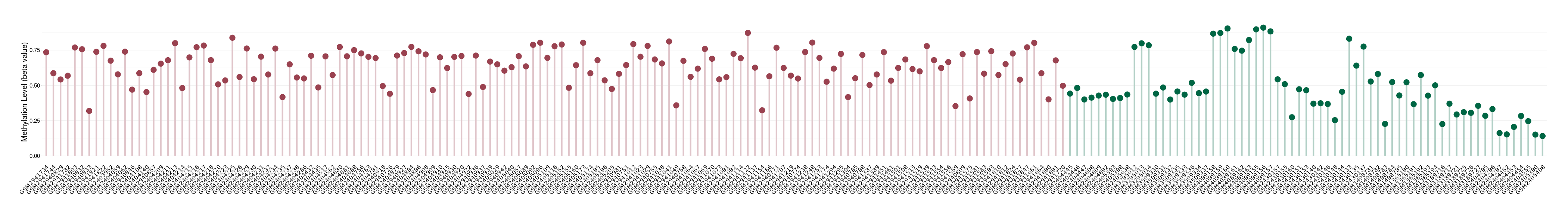 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Cerebellar liponeurocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC44A4 in cerebellar liponeurocytoma than that in healthy individual | ||||
Studied Phenotype |
Cerebellar liponeurocytoma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:5.86E-05; Fold-change:-0.293700654; Z-score:-1.539875823 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
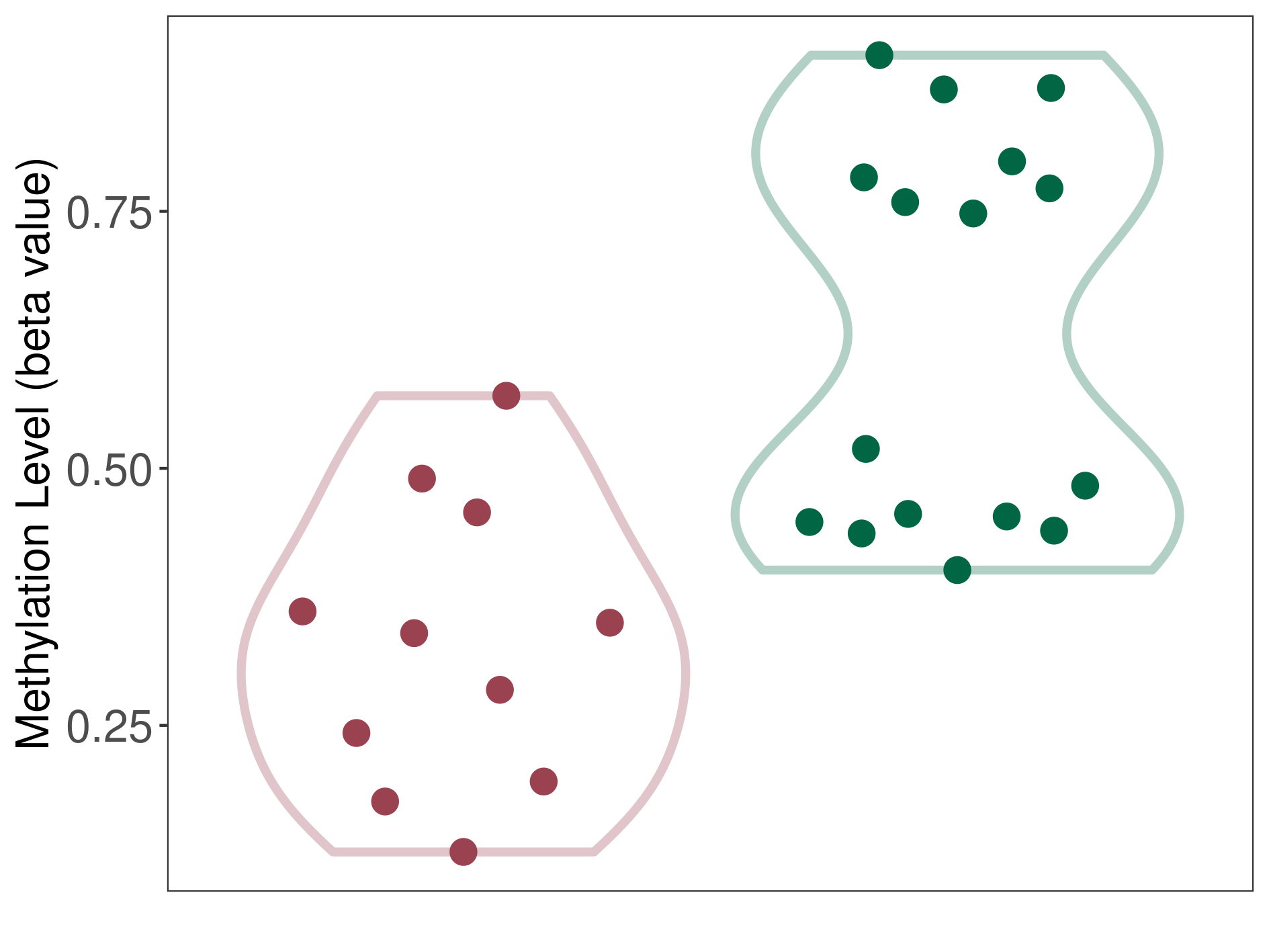
|
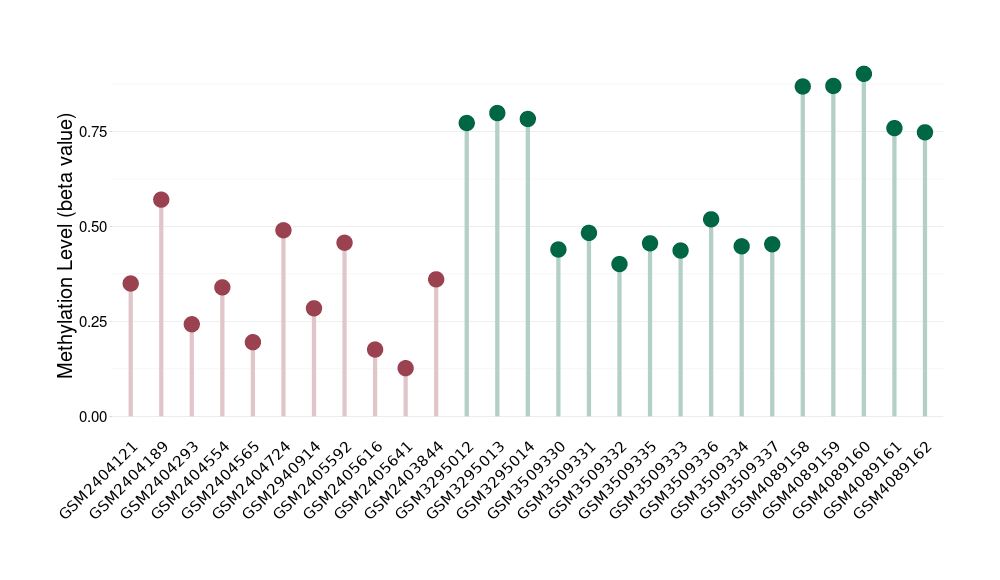 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Craniopharyngioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC44A4 in craniopharyngioma than that in healthy individual | ||||
Studied Phenotype |
Craniopharyngioma [ICD-11:2F9A] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.28E-09; Fold-change:-0.212981362; Z-score:-1.033658278 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
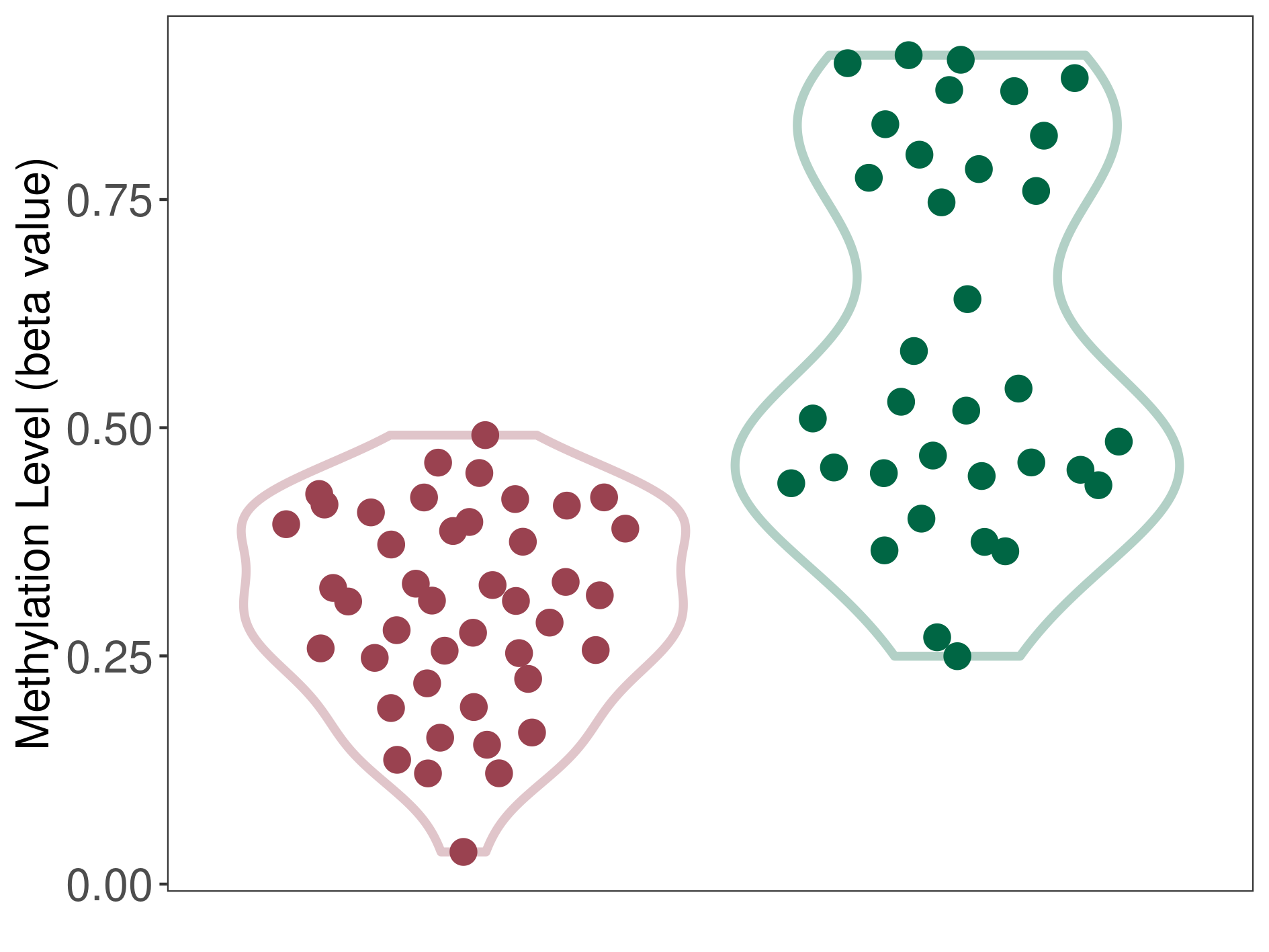
|
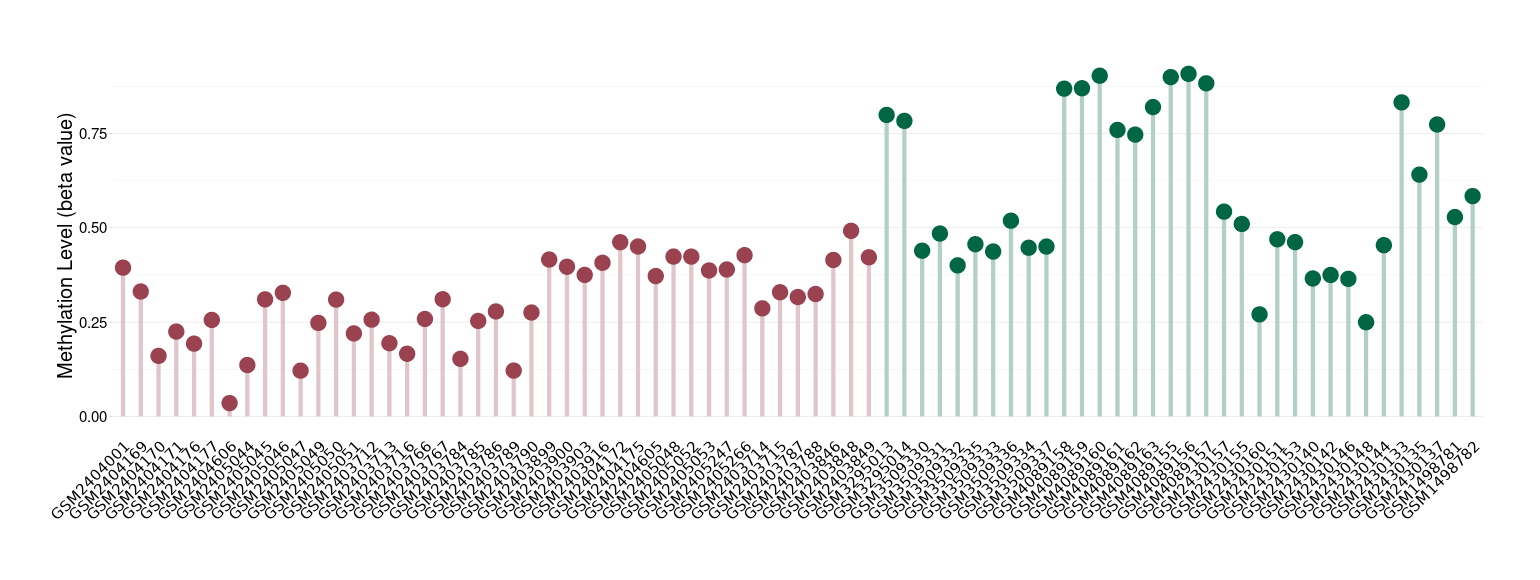 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Obesity |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC44A4 in obesity than that in healthy individual | ||||
Studied Phenotype |
Obesity [ICD-11:5B81] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:6.73E-16; Fold-change:-0.286676118; Z-score:-2.985172397 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
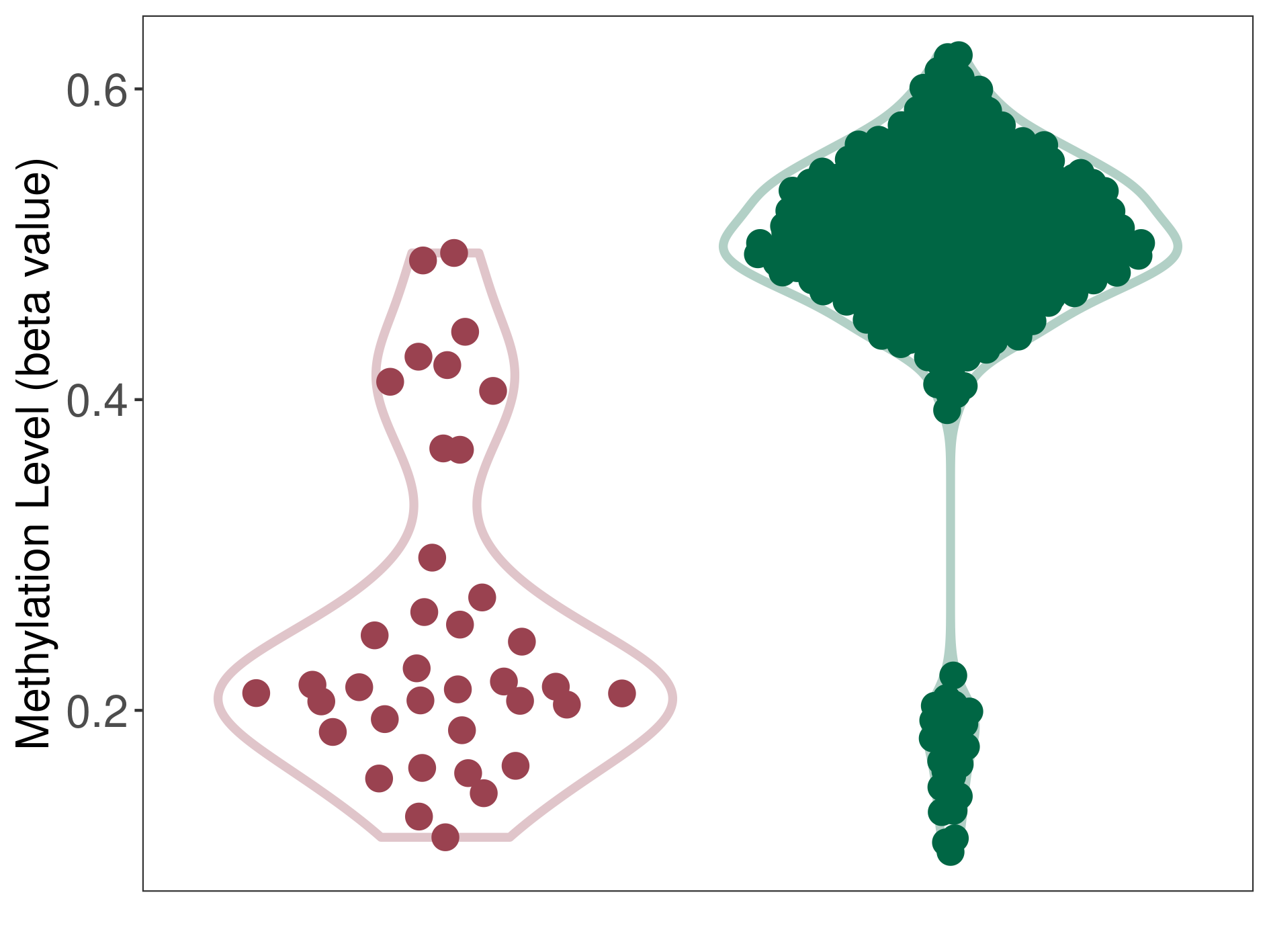
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Melanocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC44A4 in melanocytoma than that in healthy individual | ||||
Studied Phenotype |
Melanocytoma [ICD-11:2F36.2] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.035473817; Fold-change:0.321392791; Z-score:1.32584106 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
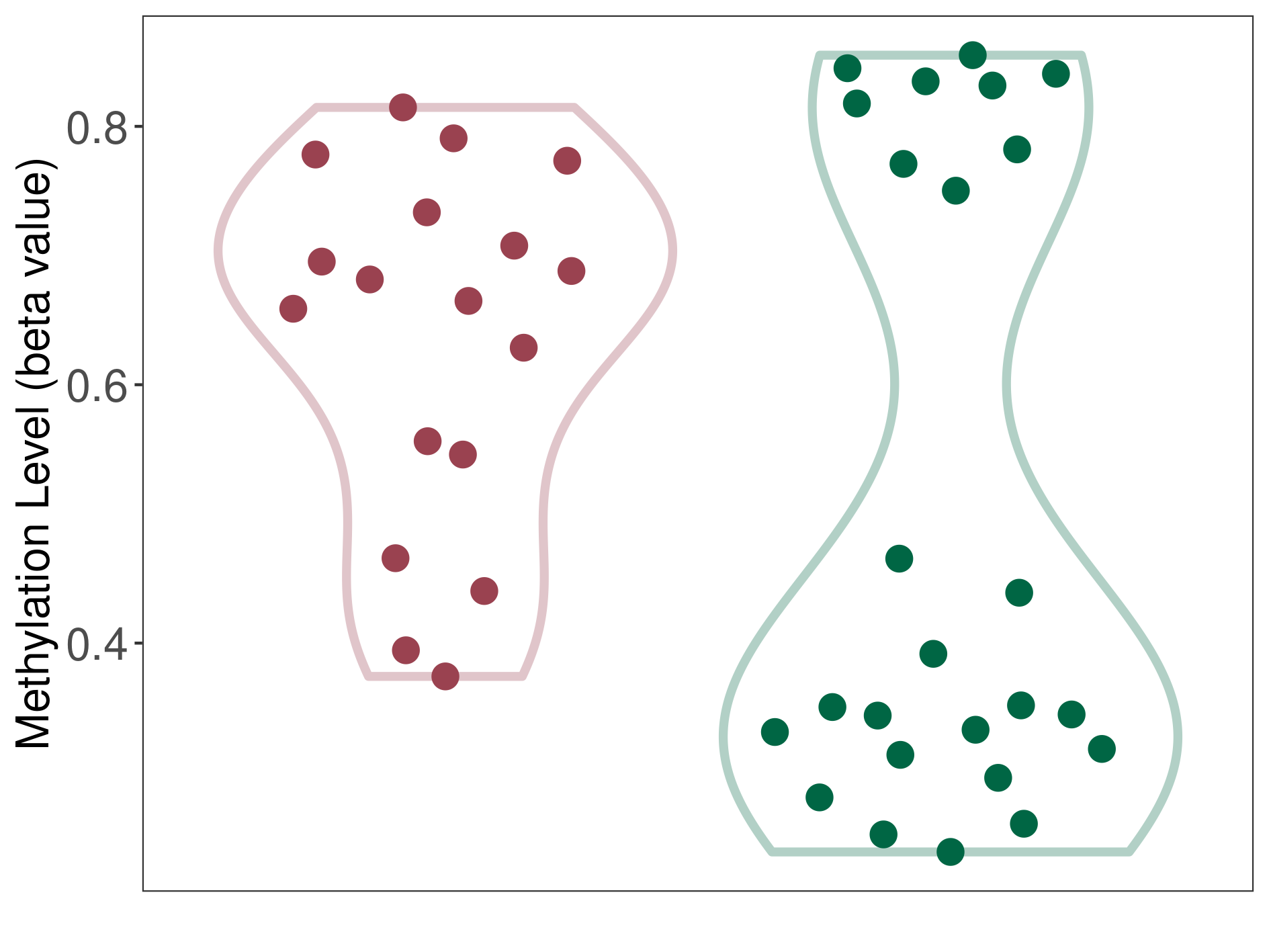
|
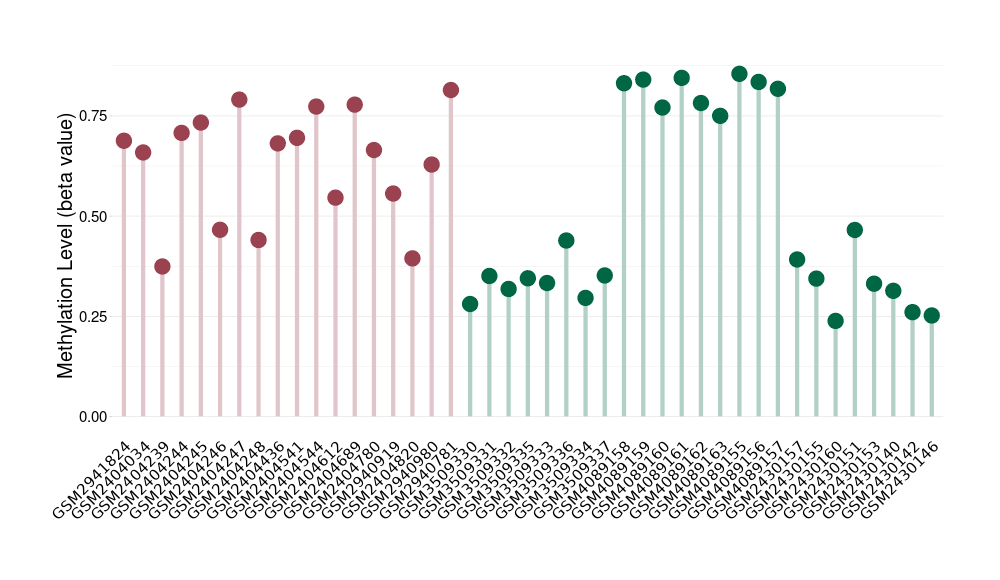 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Melanoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC44A4 in melanoma than that in healthy individual | ||||
Studied Phenotype |
Melanoma [ICD-11:2C30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.52E-11; Fold-change:0.353025708; Z-score:1.602314569 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
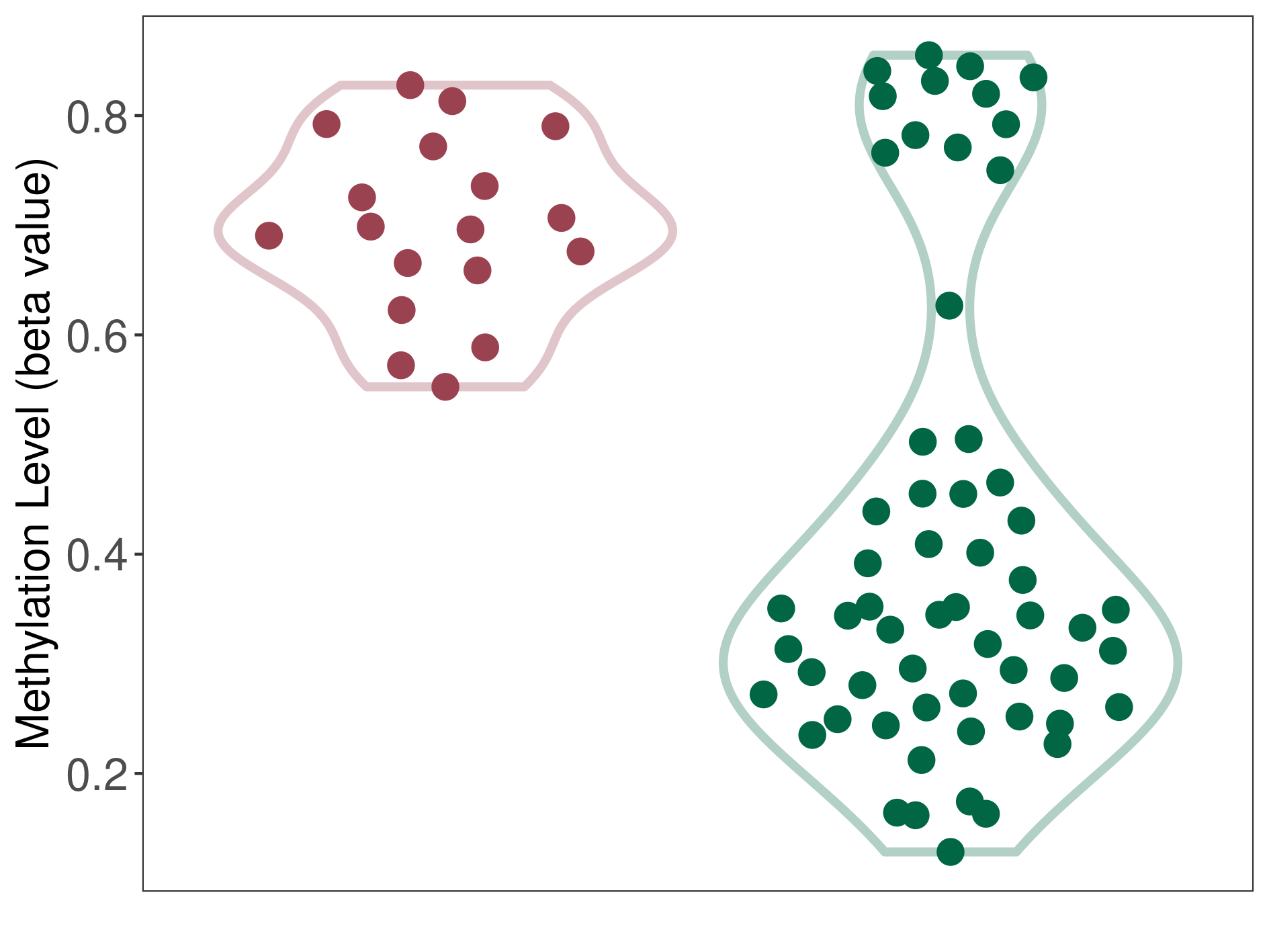
|
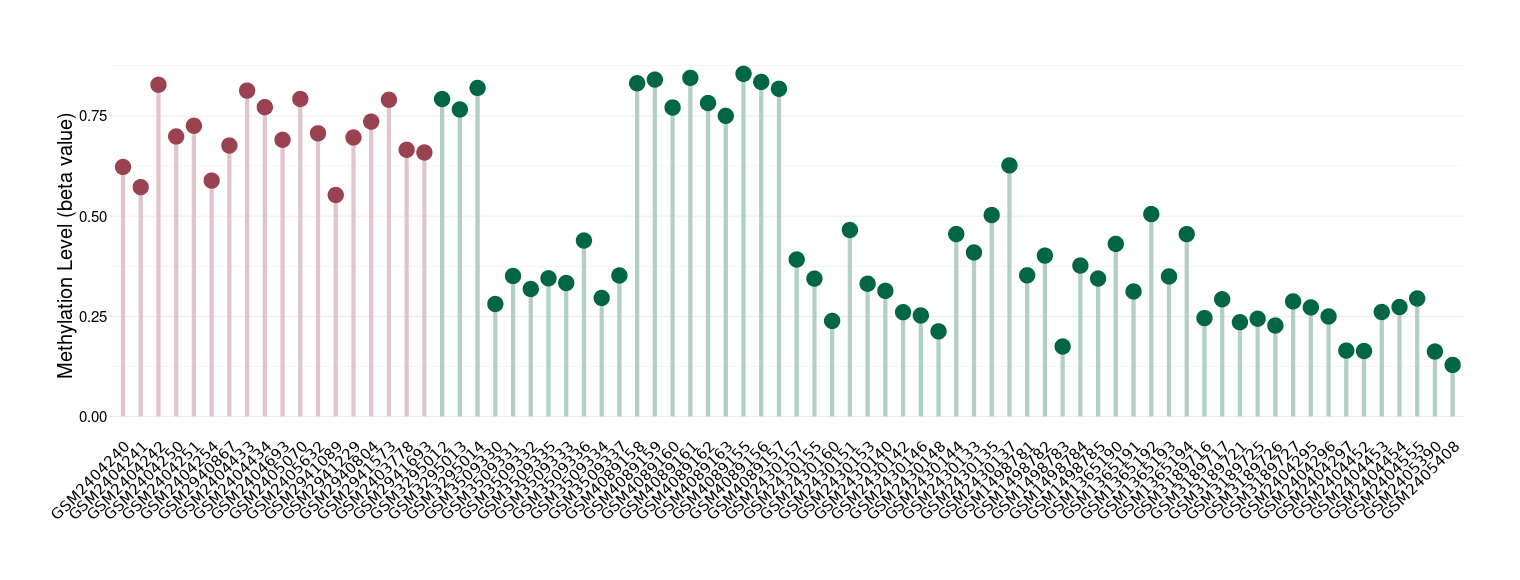 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Central neurocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC44A4 in central neurocytoma than that in healthy individual | ||||
Studied Phenotype |
Central neurocytoma [ICD-11:2A00.3] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:3.63E-06; Fold-change:-0.30719913; Z-score:-1.37094826 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
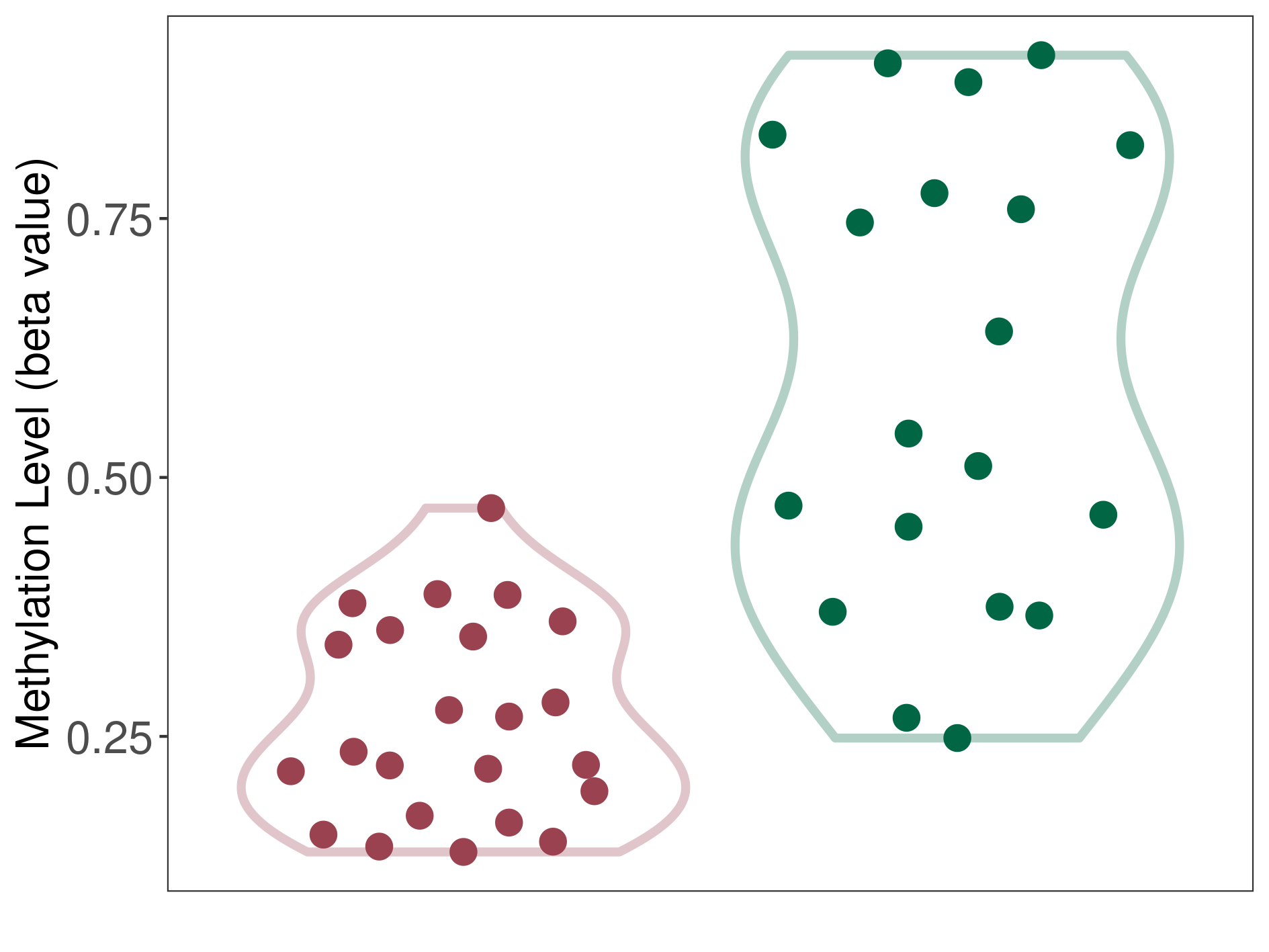
|
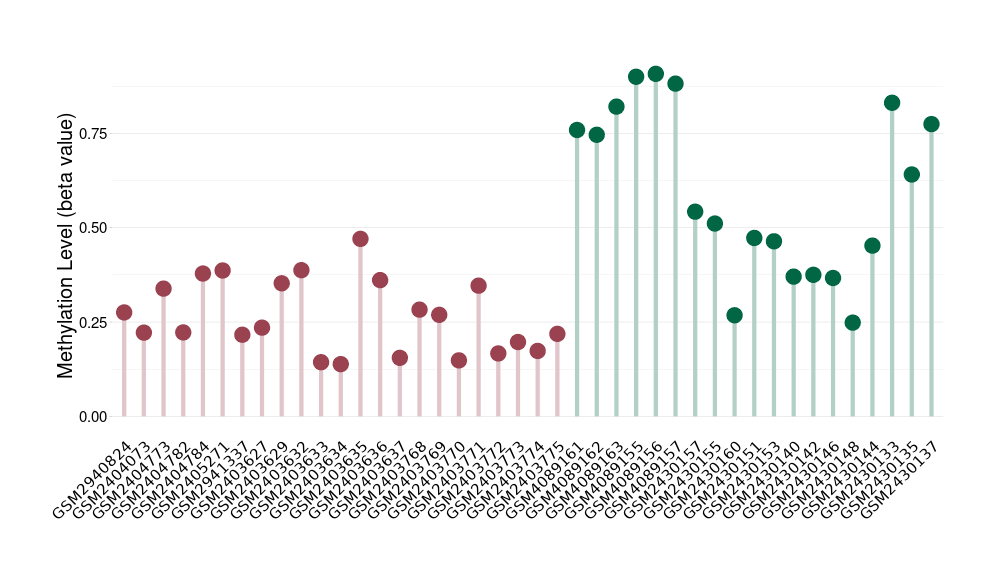 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Esthesioneuroblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC44A4 in esthesioneuroblastoma than that in healthy individual | ||||
Studied Phenotype |
Esthesioneuroblastoma [ICD-11:2D50.1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:4.98E-11; Fold-change:-0.328480899; Z-score:-1.564426545 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
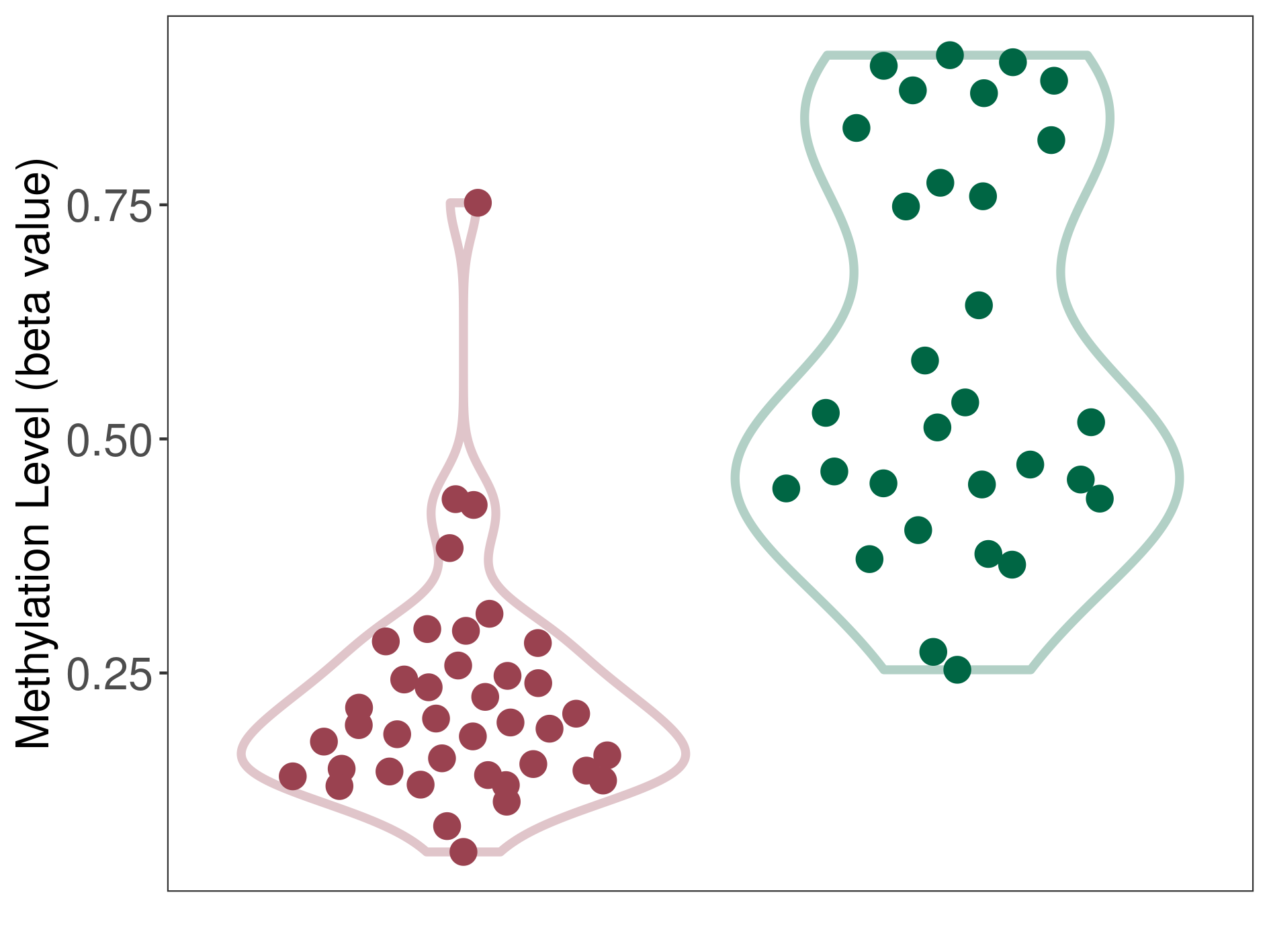
|
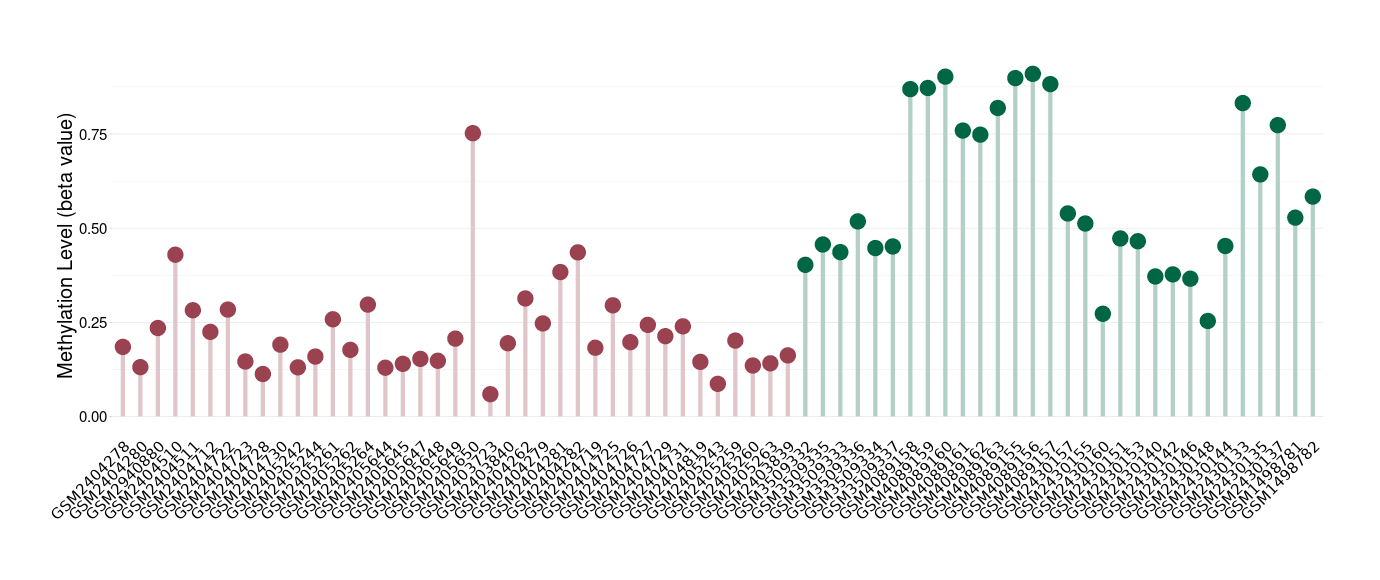 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Third ventricle chordoid glioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC44A4 in third ventricle chordoid glioma than that in healthy individual | ||||
Studied Phenotype |
Third ventricle chordoid glioma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.042335663; Fold-change:-0.302669331; Z-score:-1.176867898 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
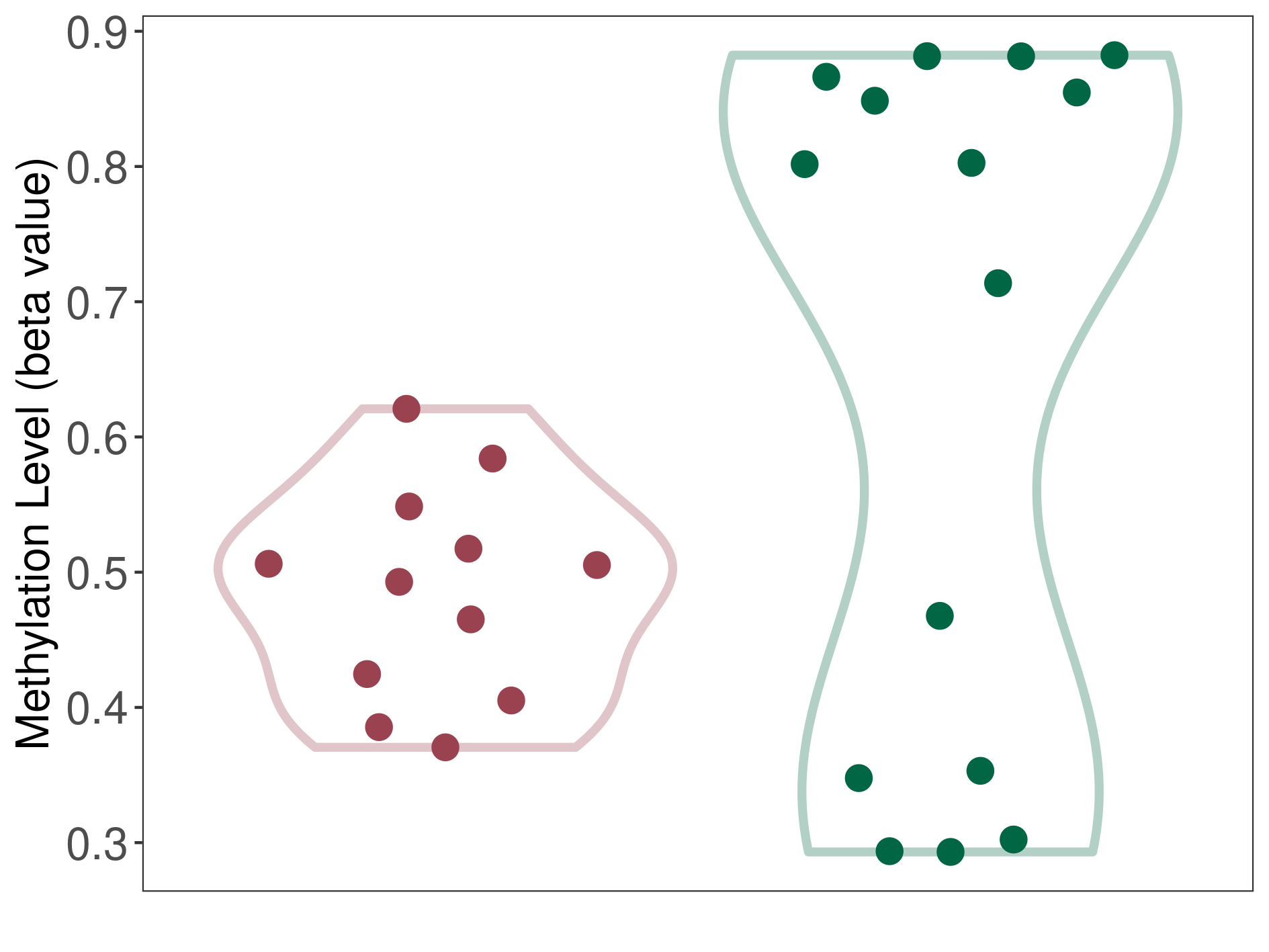
|
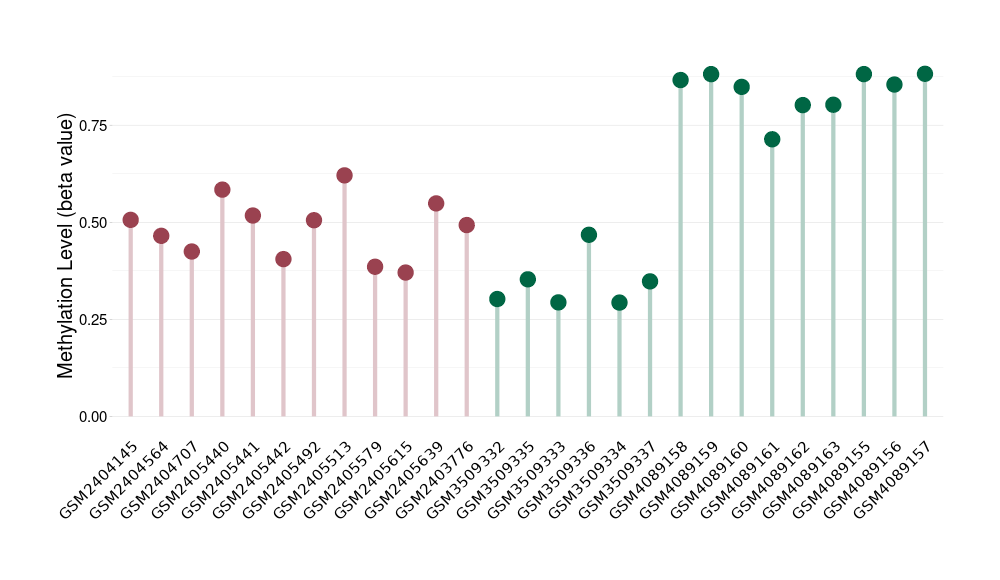 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Renal cell carcinoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC44A4 in renal cell carcinoma than that in adjacent tissue | ||||
Studied Phenotype |
Renal cell carcinoma [ICD-11:2C90] | ||||
The Methylation Level of Disease Section Compare with the Adjacent Tissue |
p-value:1.19E-05; Fold-change:-0.244441585; Z-score:-1.897247306 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
DT methylation level in tissue other than the diseased tissue of patients
|
|||||
|
microRNA |
|||||
|
Unclear Phenotype |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
miR-5187 directly targets SLC44A4 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5187 | miRNA Mature ID | miR-5187-5p | ||
|
miRNA Sequence |
UGGGAUGAGGGAUUGAAGUGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon2 |
miR-552 directly targets SLC44A4 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-552 | miRNA Mature ID | miR-552-3p | ||
|
miRNA Sequence |
AACAGGUGACUGGUUAGACAA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon3 |
miR-619 directly targets SLC44A4 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-619 | miRNA Mature ID | miR-619-5p | ||
|
miRNA Sequence |
GCUGGGAUUACAGGCAUGAGCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon4 |
miR-6506 directly targets SLC44A4 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6506 | miRNA Mature ID | miR-6506-5p | ||
|
miRNA Sequence |
ACUGGGAUGUCACUGAAUAUGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon5 |
miR-6728 directly targets SLC44A4 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6728 | miRNA Mature ID | miR-6728-5p | ||
|
miRNA Sequence |
UUGGGAUGGUAGGACCAGAGGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon6 |
miR-6744 directly targets SLC44A4 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6744 | miRNA Mature ID | miR-6744-5p | ||
|
miRNA Sequence |
UGGAUGACAGUGGAGGCCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon7 |
miR-764 directly targets SLC44A4 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-764 | miRNA Mature ID | miR-764 | ||
|
miRNA Sequence |
GCAGGUGCUCACUUGUCCUCCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
