Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0285 Transporter Info | ||||
| Gene Name | SLC34A2 | ||||
| Transporter Name | Sodium-dependent phosphate transport protein 2B | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Atypical teratoid rhabdoid tumor |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC34A2 in atypical teratoid rhabdoid tumor | [ 1 ] | |||
|
Location |
5'UTR (cg04242577) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.31E+00 | Statistic Test | p-value:2.78E-08; Z-score:1.39E+00 | ||
|
Methylation in Case |
8.43E-01 (Median) | Methylation in Control | 6.42E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC34A2 in atypical teratoid rhabdoid tumor | [ 1 ] | |||
|
Location |
5'UTR (cg19299094) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:2.75E-06; Z-score:1.09E+00 | ||
|
Methylation in Case |
8.34E-01 (Median) | Methylation in Control | 7.01E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC34A2 in atypical teratoid rhabdoid tumor | [ 1 ] | |||
|
Location |
5'UTR (cg21200703) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:4.93E-06; Z-score:-8.64E-01 | ||
|
Methylation in Case |
8.80E-01 (Median) | Methylation in Control | 9.10E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC34A2 in atypical teratoid rhabdoid tumor | [ 1 ] | |||
|
Location |
5'UTR (cg25285433) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.61E+00 | Statistic Test | p-value:9.48E-06; Z-score:1.43E+00 | ||
|
Methylation in Case |
6.93E-01 (Median) | Methylation in Control | 4.31E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC34A2 in atypical teratoid rhabdoid tumor | [ 1 ] | |||
|
Location |
5'UTR (cg25571414) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:1.06E-05; Z-score:-9.95E-01 | ||
|
Methylation in Case |
4.75E-01 (Median) | Methylation in Control | 6.46E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC34A2 in atypical teratoid rhabdoid tumor | [ 1 ] | |||
|
Location |
Body (cg01335372) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.25E+00 | Statistic Test | p-value:6.10E-05; Z-score:7.49E-01 | ||
|
Methylation in Case |
1.46E-01 (Median) | Methylation in Control | 1.16E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC34A2 in atypical teratoid rhabdoid tumor | [ 1 ] | |||
|
Location |
Body (cg06070644) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:2.20E-03; Z-score:-5.10E-01 | ||
|
Methylation in Case |
8.07E-01 (Median) | Methylation in Control | 8.33E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC34A2 in atypical teratoid rhabdoid tumor | [ 1 ] | |||
|
Location |
Body (cg13755144) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.32E+00 | Statistic Test | p-value:3.86E-02; Z-score:-7.04E-01 | ||
|
Methylation in Case |
3.07E-01 (Median) | Methylation in Control | 4.07E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC34A2 in atypical teratoid rhabdoid tumor | [ 1 ] | |||
|
Location |
3'UTR (cg03234405) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.36E+00 | Statistic Test | p-value:1.10E-14; Z-score:2.24E+00 | ||
|
Methylation in Case |
7.27E-01 (Median) | Methylation in Control | 5.35E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
8 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC34A2 in bladder cancer | [ 2 ] | |||
|
Location |
5'UTR (cg25571414) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:1.20E-03; Z-score:-3.56E+00 | ||
|
Methylation in Case |
7.67E-01 (Median) | Methylation in Control | 8.30E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC34A2 in bladder cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg02272859) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:3.97E-02; Z-score:1.75E+00 | ||
|
Methylation in Case |
7.07E-01 (Median) | Methylation in Control | 6.04E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC34A2 in bladder cancer | [ 2 ] | |||
|
Location |
Body (cg06070644) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.23E+00 | Statistic Test | p-value:8.17E-05; Z-score:-1.08E+01 | ||
|
Methylation in Case |
5.96E-01 (Median) | Methylation in Control | 7.31E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC34A2 in bladder cancer | [ 2 ] | |||
|
Location |
Body (cg13755144) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:2.88E-04; Z-score:-4.23E+00 | ||
|
Methylation in Case |
7.87E-01 (Median) | Methylation in Control | 8.89E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC34A2 in bladder cancer | [ 2 ] | |||
|
Location |
Body (cg21593669) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:2.88E-04; Z-score:-2.04E+00 | ||
|
Methylation in Case |
6.39E-01 (Median) | Methylation in Control | 6.65E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC34A2 in bladder cancer | [ 2 ] | |||
|
Location |
Body (cg01335372) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.44E-03; Z-score:-3.41E+00 | ||
|
Methylation in Case |
8.87E-01 (Median) | Methylation in Control | 9.13E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC34A2 in bladder cancer | [ 2 ] | |||
|
Location |
Body (cg16363447) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.78E-02; Z-score:-1.09E+00 | ||
|
Methylation in Case |
9.47E-01 (Median) | Methylation in Control | 9.57E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC34A2 in bladder cancer | [ 2 ] | |||
|
Location |
3'UTR (cg03234405) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.31E+00 | Statistic Test | p-value:4.16E-07; Z-score:-5.58E+00 | ||
|
Methylation in Case |
6.29E-01 (Median) | Methylation in Control | 8.23E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
19 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
5'UTR (cg19299094) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.46E+00 | Statistic Test | p-value:7.74E-10; Z-score:-2.51E+00 | ||
|
Methylation in Case |
2.41E-01 (Median) | Methylation in Control | 3.52E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
5'UTR (cg21200703) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.28E+00 | Statistic Test | p-value:1.46E-06; Z-score:1.92E+00 | ||
|
Methylation in Case |
3.24E-01 (Median) | Methylation in Control | 2.53E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
5'UTR (cg25285433) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.00E-03; Z-score:-6.29E-01 | ||
|
Methylation in Case |
6.74E-01 (Median) | Methylation in Control | 7.11E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
5'UTR (cg25571414) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.20E+00 | Statistic Test | p-value:8.06E-03; Z-score:-1.08E+00 | ||
|
Methylation in Case |
6.24E-01 (Median) | Methylation in Control | 7.48E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg02272859) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.17E+00 | Statistic Test | p-value:1.23E-08; Z-score:-2.03E+00 | ||
|
Methylation in Case |
5.52E-01 (Median) | Methylation in Control | 6.44E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg04101194) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.52E+00 | Statistic Test | p-value:3.47E-06; Z-score:-1.35E+00 | ||
|
Methylation in Case |
1.27E-01 (Median) | Methylation in Control | 1.92E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg09157302) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.28E+00 | Statistic Test | p-value:3.93E-06; Z-score:-1.04E+00 | ||
|
Methylation in Case |
5.68E-02 (Median) | Methylation in Control | 1.29E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg09752627) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.78E+00 | Statistic Test | p-value:5.60E-05; Z-score:3.65E-01 | ||
|
Methylation in Case |
4.58E-02 (Median) | Methylation in Control | 2.57E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg19766441) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.64E+00 | Statistic Test | p-value:4.35E-10; Z-score:2.62E+00 | ||
|
Methylation in Case |
1.18E-01 (Median) | Methylation in Control | 4.46E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg03738352) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.64E+00 | Statistic Test | p-value:2.19E-09; Z-score:2.02E+00 | ||
|
Methylation in Case |
1.17E-01 (Median) | Methylation in Control | 4.42E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg07578695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.76E+00 | Statistic Test | p-value:2.41E-05; Z-score:7.01E-01 | ||
|
Methylation in Case |
7.57E-02 (Median) | Methylation in Control | 4.31E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg19616230) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:6.83E-04; Z-score:4.82E-01 | ||
|
Methylation in Case |
1.96E-01 (Median) | Methylation in Control | 1.70E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg01493016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:1.83E-03; Z-score:4.60E-01 | ||
|
Methylation in Case |
1.49E-01 (Median) | Methylation in Control | 1.35E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg06070644) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:6.31E-09; Z-score:-2.02E+00 | ||
|
Methylation in Case |
6.52E-01 (Median) | Methylation in Control | 7.14E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg01335372) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.84E-06; Z-score:-1.25E+00 | ||
|
Methylation in Case |
8.80E-01 (Median) | Methylation in Control | 9.02E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg13755144) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.12E-05; Z-score:-9.67E-01 | ||
|
Methylation in Case |
8.27E-01 (Median) | Methylation in Control | 8.55E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg19241593) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.00E+00 | Statistic Test | p-value:2.41E-03; Z-score:4.08E-02 | ||
|
Methylation in Case |
8.32E-01 (Median) | Methylation in Control | 8.31E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
Body (cg16363447) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.65E-02; Z-score:-2.48E-01 | ||
|
Methylation in Case |
9.46E-01 (Median) | Methylation in Control | 9.59E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC34A2 in breast cancer | [ 3 ] | |||
|
Location |
3'UTR (cg03234405) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:5.16E-05; Z-score:-1.30E+00 | ||
|
Methylation in Case |
7.17E-01 (Median) | Methylation in Control | 7.99E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
11 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC34A2 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
5'UTR (cg21200703) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.26E+00 | Statistic Test | p-value:2.30E-04; Z-score:1.61E+00 | ||
|
Methylation in Case |
3.34E-01 (Median) | Methylation in Control | 2.65E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC34A2 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
5'UTR (cg19299094) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.17E+00 | Statistic Test | p-value:1.33E-02; Z-score:-1.71E+00 | ||
|
Methylation in Case |
3.11E-01 (Median) | Methylation in Control | 3.64E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC34A2 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
TSS1500 (cg04101194) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.43E+00 | Statistic Test | p-value:1.33E-03; Z-score:-2.40E+00 | ||
|
Methylation in Case |
2.60E-01 (Median) | Methylation in Control | 3.71E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC34A2 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
TSS1500 (cg02272859) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:2.23E-03; Z-score:-1.11E+00 | ||
|
Methylation in Case |
6.96E-01 (Median) | Methylation in Control | 7.43E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC34A2 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
TSS1500 (cg09157302) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.27E+00 | Statistic Test | p-value:4.08E-02; Z-score:-3.57E+00 | ||
|
Methylation in Case |
1.23E-01 (Median) | Methylation in Control | 2.79E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC34A2 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
TSS200 (cg03738352) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.96E+00 | Statistic Test | p-value:7.86E-05; Z-score:1.35E+00 | ||
|
Methylation in Case |
5.35E-02 (Median) | Methylation in Control | 2.73E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC34A2 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
TSS200 (cg19766441) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.88E+00 | Statistic Test | p-value:1.12E-04; Z-score:1.39E+00 | ||
|
Methylation in Case |
5.69E-02 (Median) | Methylation in Control | 3.03E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC34A2 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
TSS200 (cg19616230) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:7.73E-04; Z-score:6.66E-01 | ||
|
Methylation in Case |
1.89E-01 (Median) | Methylation in Control | 1.68E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC34A2 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
TSS200 (cg01493016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.34E+00 | Statistic Test | p-value:1.29E-03; Z-score:1.13E+00 | ||
|
Methylation in Case |
1.09E-01 (Median) | Methylation in Control | 8.12E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC34A2 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
TSS200 (cg07578695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.54E+00 | Statistic Test | p-value:1.79E-03; Z-score:9.35E-01 | ||
|
Methylation in Case |
4.55E-02 (Median) | Methylation in Control | 2.95E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC34A2 in clear cell renal cell carcinoma | [ 4 ] | |||
|
Location |
Body (cg06070644) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:5.86E-07; Z-score:-2.15E+00 | ||
|
Methylation in Case |
8.27E-01 (Median) | Methylation in Control | 8.73E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colon cancer |
10 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC34A2 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
5'UTR (cg15141217) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:2.47E-04; Z-score:-2.92E+00 | ||
|
Methylation in Case |
5.99E-01 (Median) | Methylation in Control | 6.86E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC34A2 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
5'UTR (cg24655009) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.05E-03; Z-score:-1.23E+00 | ||
|
Methylation in Case |
6.61E-01 (Median) | Methylation in Control | 6.91E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC34A2 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg18506018) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:2.79E-05; Z-score:-2.31E+00 | ||
|
Methylation in Case |
6.62E-01 (Median) | Methylation in Control | 7.67E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC34A2 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg13323701) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:4.40E+00 | Statistic Test | p-value:9.37E-07; Z-score:1.27E+01 | ||
|
Methylation in Case |
2.74E-01 (Median) | Methylation in Control | 6.23E-02 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC34A2 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg26482893) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:6.21E-05; Z-score:6.94E-01 | ||
|
Methylation in Case |
7.03E-01 (Median) | Methylation in Control | 6.18E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC34A2 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
Body (cg17057086) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:3.32E-05; Z-score:-2.29E+00 | ||
|
Methylation in Case |
5.13E-01 (Median) | Methylation in Control | 6.27E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC34A2 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
Body (cg20669572) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:1.54E-04; Z-score:1.22E+00 | ||
|
Methylation in Case |
7.74E-01 (Median) | Methylation in Control | 6.97E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC34A2 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
Body (cg03593616) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:3.19E-04; Z-score:-2.55E+00 | ||
|
Methylation in Case |
7.23E-01 (Median) | Methylation in Control | 7.92E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC34A2 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
Body (cg09183468) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:3.86E-04; Z-score:-2.54E+00 | ||
|
Methylation in Case |
7.14E-01 (Median) | Methylation in Control | 7.77E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC34A2 in colon adenocarcinoma | [ 5 ] | |||
|
Location |
3'UTR (cg09292202) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:2.43E-04; Z-score:9.59E-01 | ||
|
Methylation in Case |
7.92E-01 (Median) | Methylation in Control | 7.37E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
18 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC34A2 in colorectal cancer | [ 6 ] | |||
|
Location |
5'UTR (cg25571414) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:2.07E-05; Z-score:-1.40E+00 | ||
|
Methylation in Case |
8.53E-01 (Median) | Methylation in Control | 8.82E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC34A2 in colorectal cancer | [ 6 ] | |||
|
Location |
5'UTR (cg21200703) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:3.25E-03; Z-score:8.87E-01 | ||
|
Methylation in Case |
4.37E-01 (Median) | Methylation in Control | 3.81E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC34A2 in colorectal cancer | [ 6 ] | |||
|
Location |
5'UTR (cg19299094) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.66E-02; Z-score:-3.35E-01 | ||
|
Methylation in Case |
4.05E-01 (Median) | Methylation in Control | 4.27E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC34A2 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS1500 (cg09157302) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.68E+00 | Statistic Test | p-value:2.10E-05; Z-score:1.05E+00 | ||
|
Methylation in Case |
7.11E-02 (Median) | Methylation in Control | 4.23E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC34A2 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS1500 (cg09752627) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:3.32E-04; Z-score:1.03E-01 | ||
|
Methylation in Case |
3.41E-02 (Median) | Methylation in Control | 3.27E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC34A2 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS1500 (cg04101194) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:1.35E-03; Z-score:6.56E-01 | ||
|
Methylation in Case |
3.55E-01 (Median) | Methylation in Control | 3.09E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC34A2 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg19616230) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:6.77E-04; Z-score:7.22E-01 | ||
|
Methylation in Case |
1.52E-01 (Median) | Methylation in Control | 1.32E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC34A2 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg19766441) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:3.40E-03; Z-score:-2.38E-01 | ||
|
Methylation in Case |
5.48E-02 (Median) | Methylation in Control | 6.02E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC34A2 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg07578695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:3.88E-03; Z-score:-1.78E-01 | ||
|
Methylation in Case |
5.08E-02 (Median) | Methylation in Control | 5.41E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC34A2 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg03738352) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:4.81E-03; Z-score:-3.13E-01 | ||
|
Methylation in Case |
4.90E-02 (Median) | Methylation in Control | 5.69E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC34A2 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg01493016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:4.87E-03; Z-score:2.23E-01 | ||
|
Methylation in Case |
2.16E-01 (Median) | Methylation in Control | 2.08E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC34A2 in colorectal cancer | [ 6 ] | |||
|
Location |
Body (cg06070644) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.59E-07; Z-score:-2.27E+00 | ||
|
Methylation in Case |
8.46E-01 (Median) | Methylation in Control | 8.84E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC34A2 in colorectal cancer | [ 6 ] | |||
|
Location |
Body (cg13755144) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:3.84E-06; Z-score:-1.99E+00 | ||
|
Methylation in Case |
9.09E-01 (Median) | Methylation in Control | 9.36E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC34A2 in colorectal cancer | [ 6 ] | |||
|
Location |
Body (cg21593669) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:4.39E-06; Z-score:-1.23E+00 | ||
|
Methylation in Case |
7.04E-01 (Median) | Methylation in Control | 7.42E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC34A2 in colorectal cancer | [ 6 ] | |||
|
Location |
Body (cg01335372) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.70E-02; Z-score:-5.50E-01 | ||
|
Methylation in Case |
9.63E-01 (Median) | Methylation in Control | 9.67E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC34A2 in colorectal cancer | [ 6 ] | |||
|
Location |
Body (cg19241593) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.00E+00 | Statistic Test | p-value:2.26E-02; Z-score:3.01E-02 | ||
|
Methylation in Case |
9.43E-01 (Median) | Methylation in Control | 9.43E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC34A2 in colorectal cancer | [ 6 ] | |||
|
Location |
3'UTR (cg03234405) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:5.53E-05; Z-score:-9.71E-01 | ||
|
Methylation in Case |
8.68E-01 (Median) | Methylation in Control | 9.25E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
16 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC34A2 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
5'UTR (cg25571414) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.25E+00 | Statistic Test | p-value:8.81E-08; Z-score:-2.02E+00 | ||
|
Methylation in Case |
5.83E-01 (Median) | Methylation in Control | 7.27E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC34A2 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg26062856) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.33E+00 | Statistic Test | p-value:2.23E-10; Z-score:-2.94E+00 | ||
|
Methylation in Case |
4.07E-01 (Median) | Methylation in Control | 5.43E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC34A2 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg09752627) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.76E+00 | Statistic Test | p-value:1.63E-05; Z-score:4.41E-01 | ||
|
Methylation in Case |
7.90E-02 (Median) | Methylation in Control | 4.48E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC34A2 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg04101194) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:1.15E-04; Z-score:5.47E-01 | ||
|
Methylation in Case |
1.06E-01 (Median) | Methylation in Control | 8.80E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC34A2 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg09157302) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.51E-03; Z-score:-4.25E-02 | ||
|
Methylation in Case |
3.51E-02 (Median) | Methylation in Control | 3.65E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC34A2 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg19409156) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.31E+00 | Statistic Test | p-value:3.56E-12; Z-score:-5.44E+00 | ||
|
Methylation in Case |
6.32E-01 (Median) | Methylation in Control | 8.30E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC34A2 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg26071016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.86E+00 | Statistic Test | p-value:3.37E-10; Z-score:2.37E+00 | ||
|
Methylation in Case |
2.69E-01 (Median) | Methylation in Control | 1.45E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC34A2 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg19766441) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.37E+00 | Statistic Test | p-value:1.35E-07; Z-score:2.02E+00 | ||
|
Methylation in Case |
2.47E-01 (Median) | Methylation in Control | 1.04E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC34A2 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg03738352) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.73E+00 | Statistic Test | p-value:3.08E-06; Z-score:1.03E+00 | ||
|
Methylation in Case |
1.74E-01 (Median) | Methylation in Control | 1.00E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC34A2 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg19616230) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:1.30E-05; Z-score:1.86E-01 | ||
|
Methylation in Case |
1.72E-01 (Median) | Methylation in Control | 1.64E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC34A2 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg07578695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:3.94E-04; Z-score:1.14E-01 | ||
|
Methylation in Case |
7.93E-02 (Median) | Methylation in Control | 7.52E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC34A2 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg01493016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:7.66E-04; Z-score:1.43E-01 | ||
|
Methylation in Case |
1.48E-01 (Median) | Methylation in Control | 1.44E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC34A2 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
Body (cg09592224) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:2.56E-13; Z-score:-2.92E+00 | ||
|
Methylation in Case |
7.07E-01 (Median) | Methylation in Control | 8.02E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC34A2 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
Body (cg09130674) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.36E+00 | Statistic Test | p-value:1.62E-10; Z-score:1.58E+00 | ||
|
Methylation in Case |
5.20E-01 (Median) | Methylation in Control | 3.83E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC34A2 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
Body (cg16363447) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.11E-07; Z-score:-3.38E+00 | ||
|
Methylation in Case |
9.14E-01 (Median) | Methylation in Control | 9.65E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC34A2 in hepatocellular carcinoma | [ 7 ] | |||
|
Location |
Body (cg01335372) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.07E-05; Z-score:-4.09E-01 | ||
|
Methylation in Case |
8.55E-01 (Median) | Methylation in Control | 9.01E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
HIV infection |
13 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC34A2 in HIV infection | [ 8 ] | |||
|
Location |
5'UTR (cg21200703) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:8.72E-06; Z-score:1.35E+00 | ||
|
Methylation in Case |
4.46E-01 (Median) | Methylation in Control | 3.65E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC34A2 in HIV infection | [ 8 ] | |||
|
Location |
5'UTR (cg19299094) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:3.03E-04; Z-score:-5.67E-01 | ||
|
Methylation in Case |
2.33E-01 (Median) | Methylation in Control | 2.61E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC34A2 in HIV infection | [ 8 ] | |||
|
Location |
5'UTR (cg25571414) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:4.89E-02; Z-score:-6.35E-01 | ||
|
Methylation in Case |
8.53E-01 (Median) | Methylation in Control | 8.75E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC34A2 in HIV infection | [ 8 ] | |||
|
Location |
TSS1500 (cg09752627) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.45E+00 | Statistic Test | p-value:1.38E-04; Z-score:3.08E+00 | ||
|
Methylation in Case |
7.18E-02 (Median) | Methylation in Control | 2.93E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC34A2 in HIV infection | [ 8 ] | |||
|
Location |
TSS1500 (cg09157302) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.52E+00 | Statistic Test | p-value:2.63E-04; Z-score:1.10E+00 | ||
|
Methylation in Case |
2.35E-02 (Median) | Methylation in Control | 1.55E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC34A2 in HIV infection | [ 8 ] | |||
|
Location |
TSS1500 (cg04101194) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:1.72E-03; Z-score:9.04E-01 | ||
|
Methylation in Case |
6.66E-02 (Median) | Methylation in Control | 5.45E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC34A2 in HIV infection | [ 8 ] | |||
|
Location |
TSS200 (cg19616230) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.35E+00 | Statistic Test | p-value:1.04E-05; Z-score:2.33E+00 | ||
|
Methylation in Case |
2.48E-01 (Median) | Methylation in Control | 1.84E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC34A2 in HIV infection | [ 8 ] | |||
|
Location |
TSS200 (cg07578695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.07E+00 | Statistic Test | p-value:3.30E-05; Z-score:2.67E+00 | ||
|
Methylation in Case |
1.25E-01 (Median) | Methylation in Control | 6.00E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC34A2 in HIV infection | [ 8 ] | |||
|
Location |
TSS200 (cg01493016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.32E+00 | Statistic Test | p-value:3.65E-05; Z-score:1.86E+00 | ||
|
Methylation in Case |
1.79E-01 (Median) | Methylation in Control | 1.36E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC34A2 in HIV infection | [ 8 ] | |||
|
Location |
TSS200 (cg19766441) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.31E+00 | Statistic Test | p-value:4.65E-05; Z-score:3.12E+00 | ||
|
Methylation in Case |
1.31E-01 (Median) | Methylation in Control | 5.67E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC34A2 in HIV infection | [ 8 ] | |||
|
Location |
TSS200 (cg03738352) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.94E+00 | Statistic Test | p-value:6.56E-05; Z-score:2.23E+00 | ||
|
Methylation in Case |
1.49E-01 (Median) | Methylation in Control | 7.68E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC34A2 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg13755144) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.07E-02; Z-score:-2.47E-01 | ||
|
Methylation in Case |
9.24E-01 (Median) | Methylation in Control | 9.29E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC34A2 in HIV infection | [ 8 ] | |||
|
Location |
Body (cg06070644) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.36E-02; Z-score:-3.36E-01 | ||
|
Methylation in Case |
7.58E-01 (Median) | Methylation in Control | 7.69E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Pancretic ductal adenocarcinoma |
14 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC34A2 in pancretic ductal adenocarcinoma | [ 9 ] | |||
|
Location |
5'UTR (cg03553947) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.95E-07; Z-score:-6.66E-01 | ||
|
Methylation in Case |
7.21E-01 (Median) | Methylation in Control | 7.42E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC34A2 in pancretic ductal adenocarcinoma | [ 9 ] | |||
|
Location |
5'UTR (cg12929567) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:3.23E-05; Z-score:3.99E-01 | ||
|
Methylation in Case |
2.53E-02 (Median) | Methylation in Control | 2.24E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC34A2 in pancretic ductal adenocarcinoma | [ 9 ] | |||
|
Location |
TSS1500 (cg01770362) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:9.97E-11; Z-score:-2.28E+00 | ||
|
Methylation in Case |
5.31E-01 (Median) | Methylation in Control | 6.00E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC34A2 in pancretic ductal adenocarcinoma | [ 9 ] | |||
|
Location |
TSS1500 (cg01557989) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.24E-07; Z-score:-1.55E-01 | ||
|
Methylation in Case |
8.92E-02 (Median) | Methylation in Control | 9.39E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC34A2 in pancretic ductal adenocarcinoma | [ 9 ] | |||
|
Location |
TSS1500 (cg21220147) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:1.63E-04; Z-score:-5.91E-01 | ||
|
Methylation in Case |
1.09E-01 (Median) | Methylation in Control | 1.25E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC34A2 in pancretic ductal adenocarcinoma | [ 9 ] | |||
|
Location |
TSS1500 (cg27382790) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:1.57E-03; Z-score:-8.80E-01 | ||
|
Methylation in Case |
1.77E-01 (Median) | Methylation in Control | 2.06E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC34A2 in pancretic ductal adenocarcinoma | [ 9 ] | |||
|
Location |
TSS200 (cg16644457) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.66E+00 | Statistic Test | p-value:1.17E-12; Z-score:2.05E+00 | ||
|
Methylation in Case |
2.30E-01 (Median) | Methylation in Control | 1.39E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC34A2 in pancretic ductal adenocarcinoma | [ 9 ] | |||
|
Location |
TSS200 (cg23715104) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:2.61E-04; Z-score:1.05E+00 | ||
|
Methylation in Case |
7.10E-01 (Median) | Methylation in Control | 6.33E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC34A2 in pancretic ductal adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg03139228) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:4.00E-13; Z-score:-2.09E+00 | ||
|
Methylation in Case |
8.28E-01 (Median) | Methylation in Control | 8.92E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC34A2 in pancretic ductal adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg14486338) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.60E+00 | Statistic Test | p-value:5.65E-11; Z-score:2.01E+00 | ||
|
Methylation in Case |
5.65E-01 (Median) | Methylation in Control | 3.53E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC34A2 in pancretic ductal adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg10601171) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:2.77E-09; Z-score:1.80E+00 | ||
|
Methylation in Case |
7.21E-01 (Median) | Methylation in Control | 6.63E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC34A2 in pancretic ductal adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg12848639) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:2.90E-08; Z-score:1.50E+00 | ||
|
Methylation in Case |
7.48E-01 (Median) | Methylation in Control | 6.81E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC34A2 in pancretic ductal adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg15527515) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:2.00E-04; Z-score:-9.25E-01 | ||
|
Methylation in Case |
7.31E-01 (Median) | Methylation in Control | 8.00E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC34A2 in pancretic ductal adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg18534312) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:4.84E-03; Z-score:-4.00E-01 | ||
|
Methylation in Case |
6.20E-01 (Median) | Methylation in Control | 6.68E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC34A2 in panic disorder | [ 10 ] | |||
|
Location |
5'UTR (cg25571414) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:5.78E-03; Z-score:8.05E-01 | ||
|
Methylation in Case |
2.25E+00 (Median) | Methylation in Control | 2.03E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
13 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC34A2 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
5'UTR (cg25285433) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.49E+00 | Statistic Test | p-value:9.32E-21; Z-score:-7.77E+00 | ||
|
Methylation in Case |
5.62E-01 (Median) | Methylation in Control | 8.35E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC34A2 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
5'UTR (cg25571414) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.37E+00 | Statistic Test | p-value:4.14E-18; Z-score:-6.71E+00 | ||
|
Methylation in Case |
6.48E-01 (Median) | Methylation in Control | 8.86E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC34A2 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
5'UTR (cg19299094) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.62E+00 | Statistic Test | p-value:3.84E-16; Z-score:-2.42E+00 | ||
|
Methylation in Case |
1.99E-01 (Median) | Methylation in Control | 3.23E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC34A2 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
5'UTR (cg04242577) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.41E+00 | Statistic Test | p-value:7.48E-14; Z-score:-2.91E+00 | ||
|
Methylation in Case |
3.84E-01 (Median) | Methylation in Control | 5.41E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC34A2 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
TSS1500 (cg04101194) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.73E+00 | Statistic Test | p-value:5.23E-11; Z-score:-2.02E+00 | ||
|
Methylation in Case |
2.36E-01 (Median) | Methylation in Control | 4.08E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC34A2 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
TSS1500 (cg02272859) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:5.60E-08; Z-score:-2.03E+00 | ||
|
Methylation in Case |
5.72E-01 (Median) | Methylation in Control | 6.99E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC34A2 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
TSS1500 (cg09157302) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.56E+00 | Statistic Test | p-value:7.29E-05; Z-score:-1.41E+00 | ||
|
Methylation in Case |
2.35E-01 (Median) | Methylation in Control | 3.67E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC34A2 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
TSS200 (cg19616230) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.23E+00 | Statistic Test | p-value:1.52E-11; Z-score:2.12E+00 | ||
|
Methylation in Case |
2.35E-01 (Median) | Methylation in Control | 1.91E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC34A2 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
TSS200 (cg01493016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.28E+00 | Statistic Test | p-value:5.45E-11; Z-score:2.20E+00 | ||
|
Methylation in Case |
1.40E-01 (Median) | Methylation in Control | 1.10E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC34A2 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
TSS200 (cg07578695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.42E+00 | Statistic Test | p-value:1.63E-08; Z-score:1.62E+00 | ||
|
Methylation in Case |
1.13E-01 (Median) | Methylation in Control | 7.98E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC34A2 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
TSS200 (cg03738352) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.25E+00 | Statistic Test | p-value:1.37E-06; Z-score:9.45E-01 | ||
|
Methylation in Case |
8.59E-02 (Median) | Methylation in Control | 6.88E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC34A2 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
TSS200 (cg19766441) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.26E+00 | Statistic Test | p-value:3.82E-04; Z-score:8.59E-01 | ||
|
Methylation in Case |
8.04E-02 (Median) | Methylation in Control | 6.39E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC34A2 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg06070644) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:2.57E-02; Z-score:-7.08E-02 | ||
|
Methylation in Case |
8.29E-01 (Median) | Methylation in Control | 8.31E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC34A2 in prostate cancer | [ 12 ] | |||
|
Location |
5'UTR (cg14957718) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:4.62E+00 | Statistic Test | p-value:6.10E-04; Z-score:4.94E+00 | ||
|
Methylation in Case |
5.50E-01 (Median) | Methylation in Control | 1.19E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC34A2 in prostate cancer | [ 12 ] | |||
|
Location |
TSS1500 (cg05152717) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:2.72E-02; Z-score:1.39E+00 | ||
|
Methylation in Case |
7.48E-02 (Median) | Methylation in Control | 6.19E-02 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC34A2 in prostate cancer | [ 12 ] | |||
|
Location |
TSS1500 (cg17649104) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:2.92E-02; Z-score:1.79E+00 | ||
|
Methylation in Case |
9.62E-01 (Median) | Methylation in Control | 9.45E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC34A2 in prostate cancer | [ 12 ] | |||
|
Location |
TSS1500 (cg04422990) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.61E+00 | Statistic Test | p-value:3.29E-02; Z-score:5.47E+00 | ||
|
Methylation in Case |
5.43E-01 (Median) | Methylation in Control | 3.38E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC34A2 in prostate cancer | [ 12 ] | |||
|
Location |
Body (cg02124920) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:3.80E-02; Z-score:2.01E+00 | ||
|
Methylation in Case |
8.45E-01 (Median) | Methylation in Control | 7.40E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC34A2 in prostate cancer | [ 12 ] | |||
|
Location |
Body (cg02390715) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:4.09E-02; Z-score:1.40E+00 | ||
|
Methylation in Case |
8.79E-01 (Median) | Methylation in Control | 7.96E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Significant hypermethylation of SLC34A2 in prostate cancer than that in healthy individual | ||||
Studied Phenotype |
Prostate cancer [ICD-11:2C82] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.53E-55; Fold-change:0.321665716; Z-score:16.83774832 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
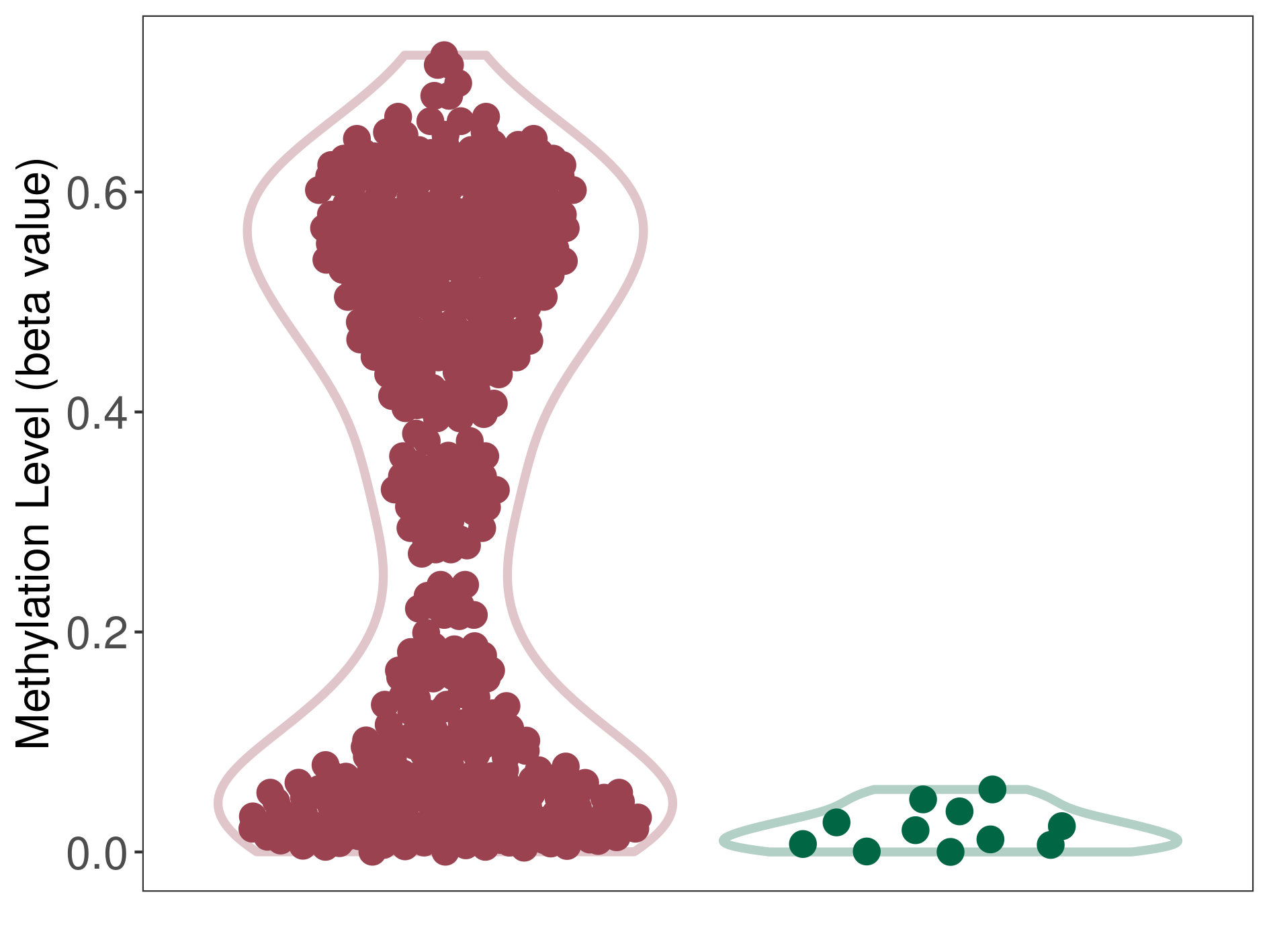
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Systemic lupus erythematosus |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC34A2 in systemic lupus erythematosus | [ 13 ] | |||
|
Location |
5'UTR (cg19299094) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:2.42E-02; Z-score:-1.01E-01 | ||
|
Methylation in Case |
2.92E-01 (Median) | Methylation in Control | 2.99E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC34A2 in systemic lupus erythematosus | [ 13 ] | |||
|
Location |
TSS200 (cg03738352) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:9.25E-03; Z-score:2.40E-01 | ||
|
Methylation in Case |
1.05E-01 (Median) | Methylation in Control | 9.34E-02 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC34A2 in systemic lupus erythematosus | [ 13 ] | |||
|
Location |
Body (cg13755144) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.53E-03; Z-score:-3.21E-01 | ||
|
Methylation in Case |
9.06E-01 (Median) | Methylation in Control | 9.14E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC34A2 in systemic lupus erythematosus | [ 13 ] | |||
|
Location |
Body (cg01335372) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.95E-02; Z-score:-3.09E-01 | ||
|
Methylation in Case |
9.30E-01 (Median) | Methylation in Control | 9.35E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC34A2 in systemic lupus erythematosus | [ 13 ] | |||
|
Location |
3'UTR (cg03234405) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.93E-02; Z-score:-6.59E-02 | ||
|
Methylation in Case |
8.78E-01 (Median) | Methylation in Control | 8.80E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC34A2 in lung adenocarcinoma | [ 14 ] | |||
|
Location |
TSS1500 (cg09752627) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.64E+00 | Statistic Test | p-value:4.23E-02; Z-score:2.46E+00 | ||
|
Methylation in Case |
9.55E-02 (Median) | Methylation in Control | 5.83E-02 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC34A2 in lung adenocarcinoma | [ 14 ] | |||
|
Location |
TSS200 (cg07578695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.58E+00 | Statistic Test | p-value:1.83E-02; Z-score:1.95E+00 | ||
|
Methylation in Case |
1.40E-01 (Median) | Methylation in Control | 8.84E-02 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC34A2 in lung adenocarcinoma | [ 14 ] | |||
|
Location |
TSS200 (cg19616230) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:2.53E-02; Z-score:2.22E+00 | ||
|
Methylation in Case |
2.62E-01 (Median) | Methylation in Control | 2.15E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC34A2 in lung adenocarcinoma | [ 14 ] | |||
|
Location |
TSS200 (cg03738352) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.63E+00 | Statistic Test | p-value:3.87E-02; Z-score:1.82E+00 | ||
|
Methylation in Case |
1.32E-01 (Median) | Methylation in Control | 8.07E-02 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC34A2 in lung adenocarcinoma | [ 14 ] | |||
|
Location |
TSS200 (cg19766441) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.54E+00 | Statistic Test | p-value:4.06E-02; Z-score:2.21E+00 | ||
|
Methylation in Case |
1.38E-01 (Median) | Methylation in Control | 8.99E-02 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Depression |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC34A2 in depression | [ 15 ] | |||
|
Location |
TSS200 (cg01493016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:4.32E-02; Z-score:3.49E-01 | ||
|
Methylation in Case |
1.09E-01 (Median) | Methylation in Control | 1.05E-01 (Median) | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Anaplastic pleomorphic xanthoastrocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC34A2 in anaplastic pleomorphic xanthoastrocytoma than that in healthy individual | ||||
Studied Phenotype |
Anaplastic pleomorphic xanthoastrocytoma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.000428175; Fold-change:0.414241969; Z-score:1.539133419 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
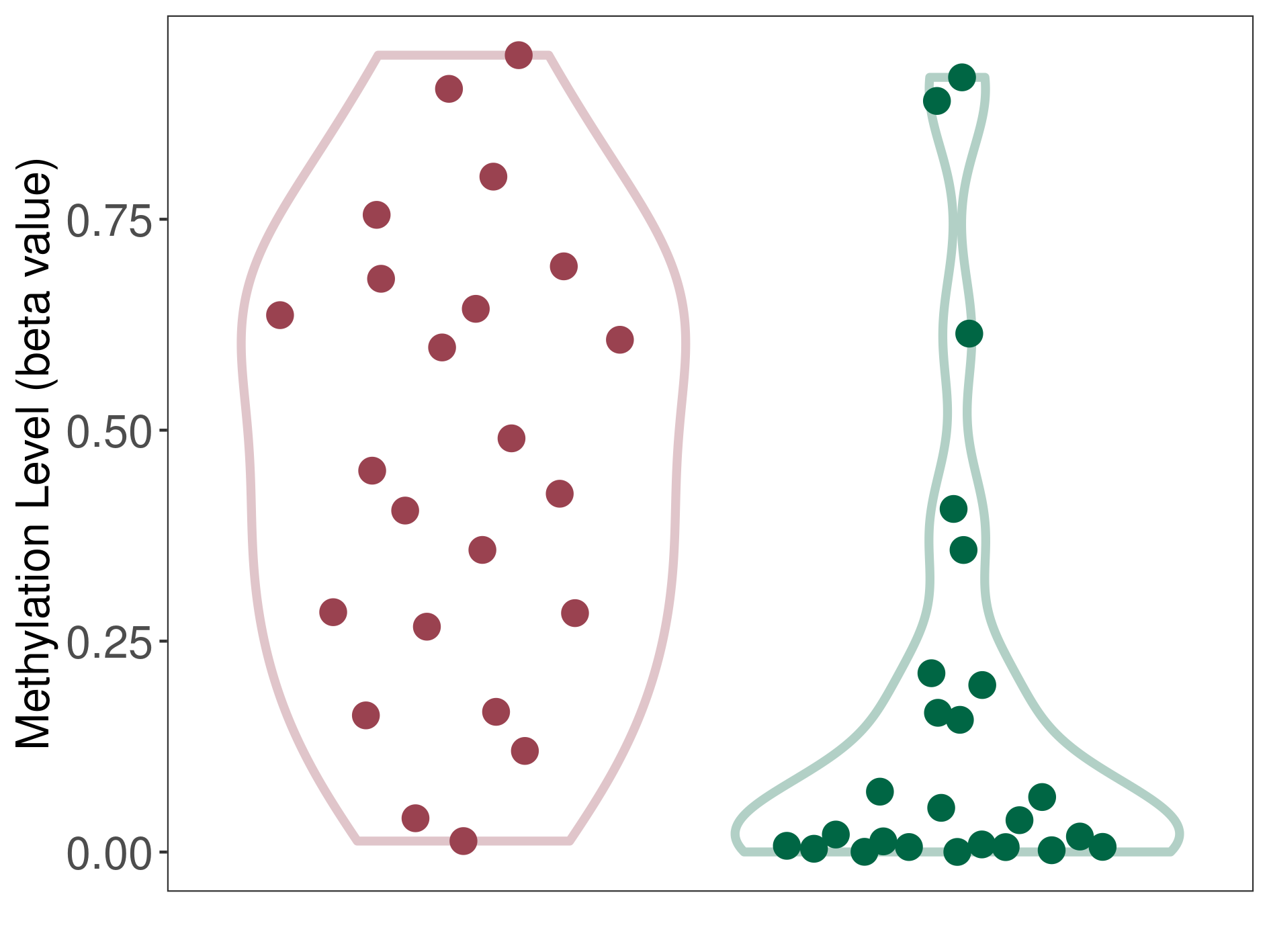
|
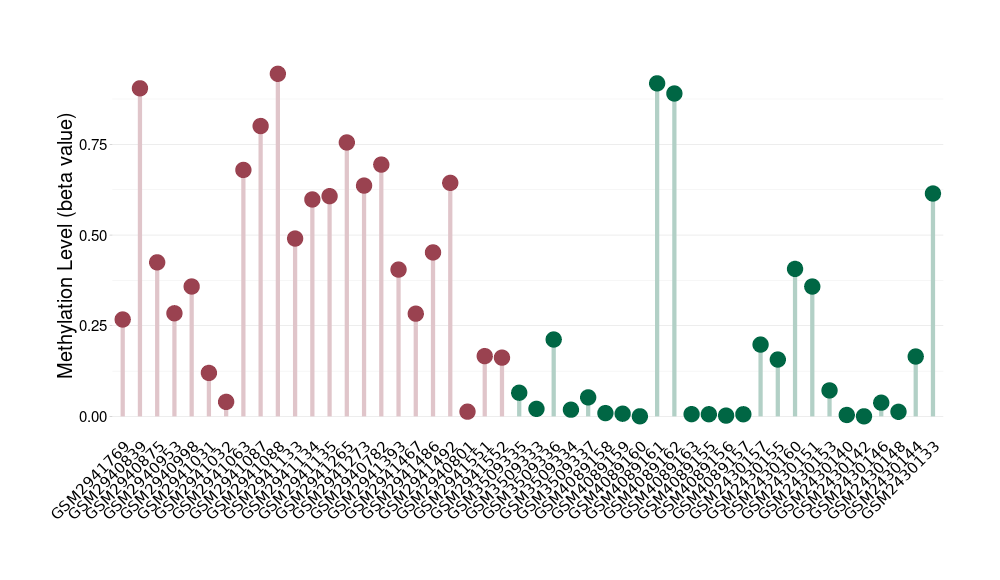 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Atypical teratoid rhabdoid tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC34A2 in atypical teratoid rhabdoid tumour than that in healthy individual | ||||
Studied Phenotype |
Atypical teratoid rhabdoid tumour [ICD-11:2A00.1Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:4.51E-05; Fold-change:0.43144968; Z-score:1.314688365 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
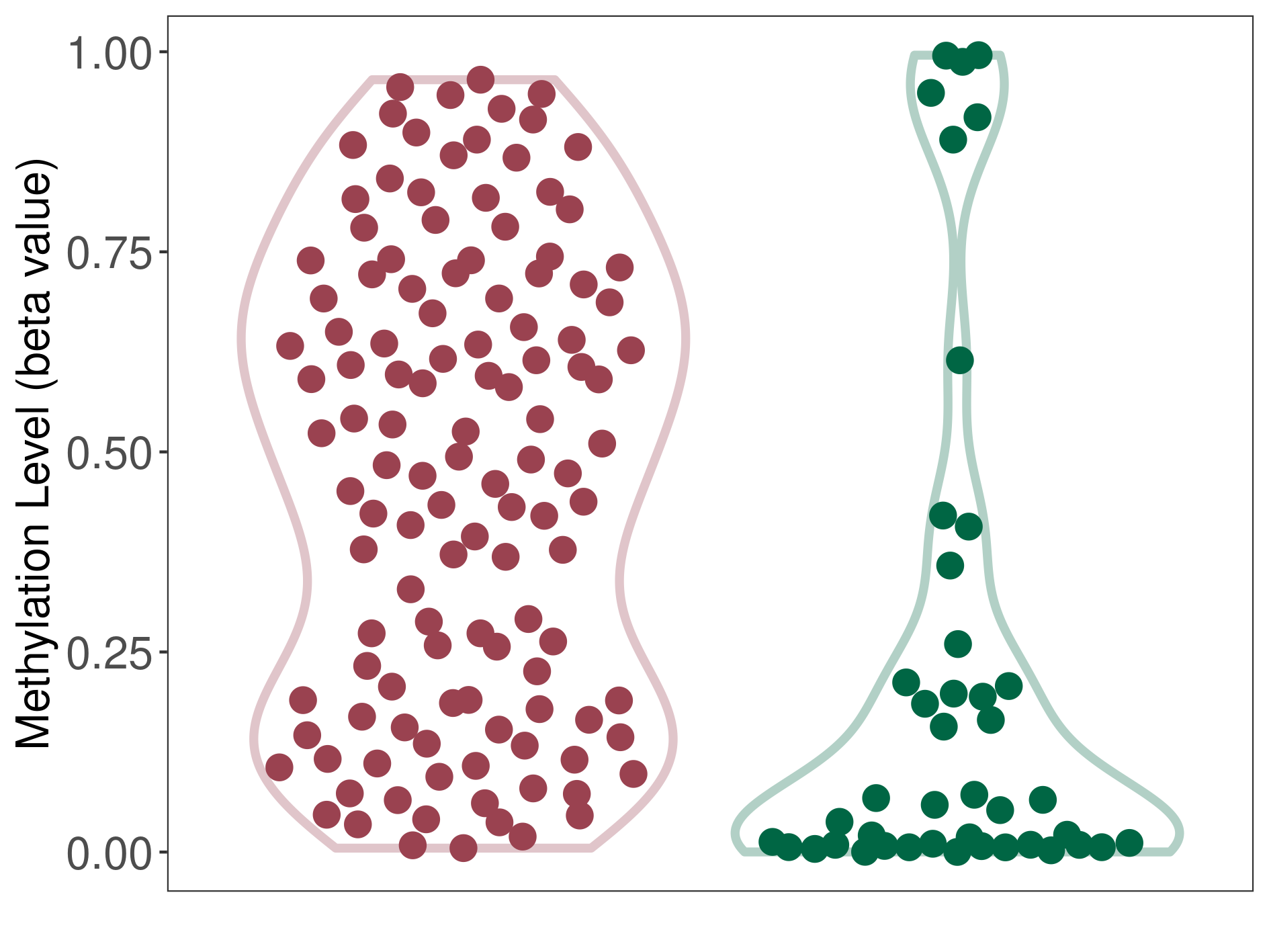
|
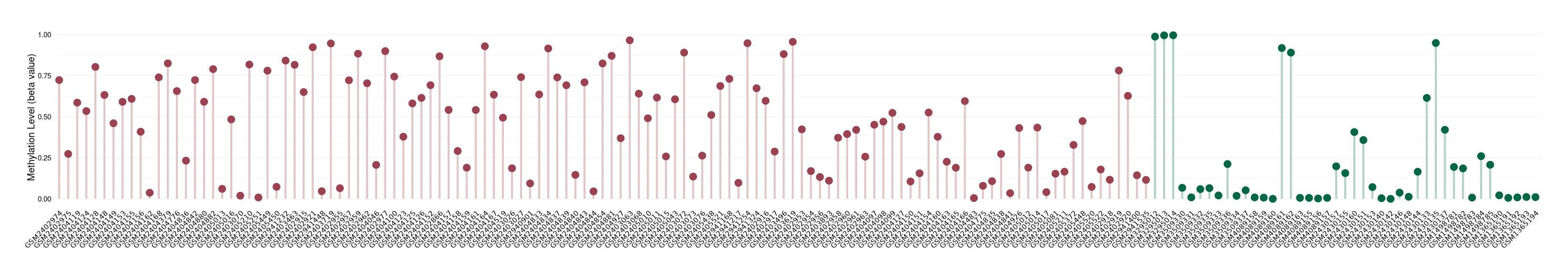 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Brain neuroblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC34A2 in brain neuroblastoma than that in healthy individual | ||||
Studied Phenotype |
Brain neuroblastoma [ICD-11:2A00.11] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:8.20E-11; Fold-change:0.586712242; Z-score:2.239686466 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
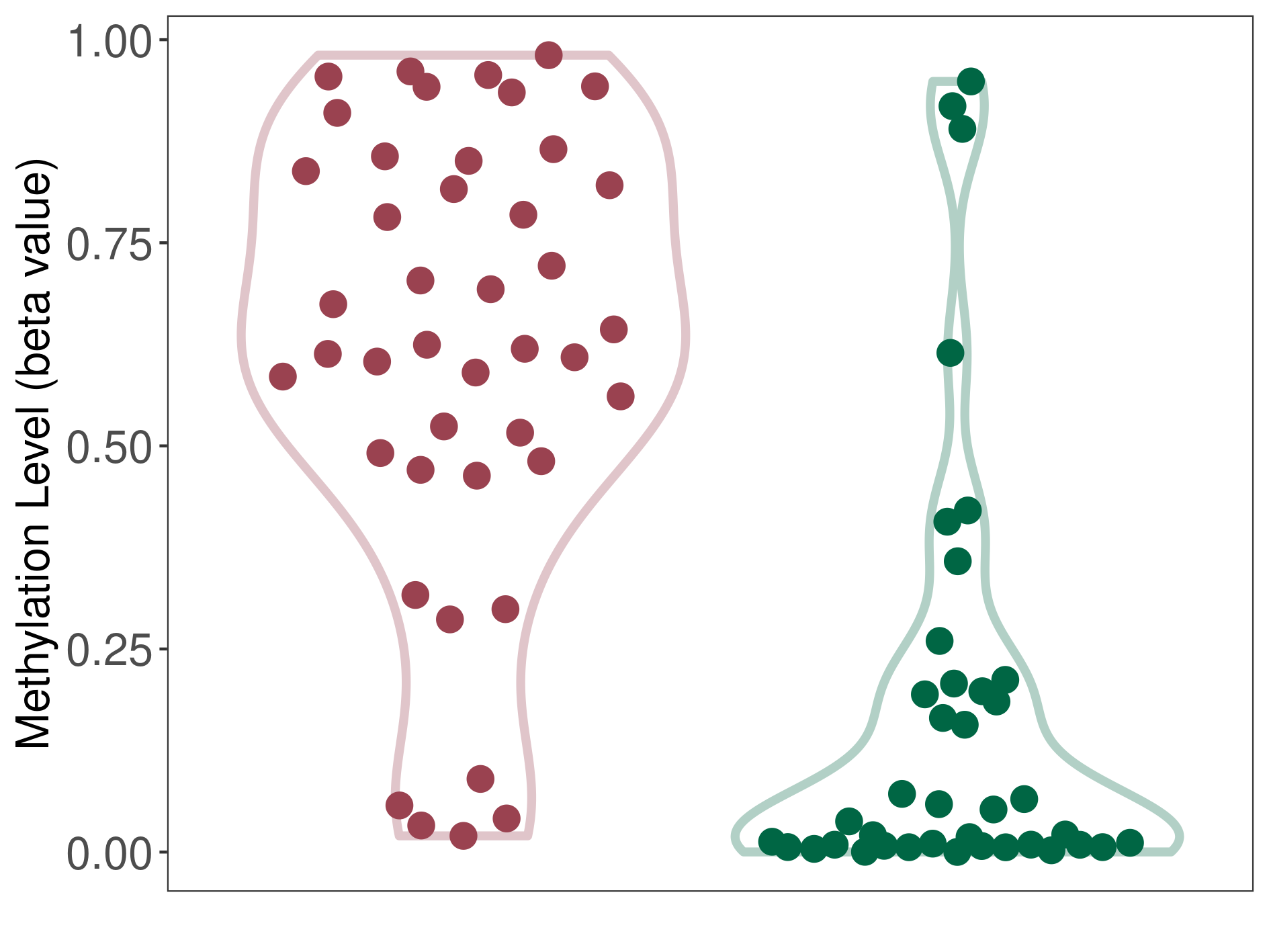
|
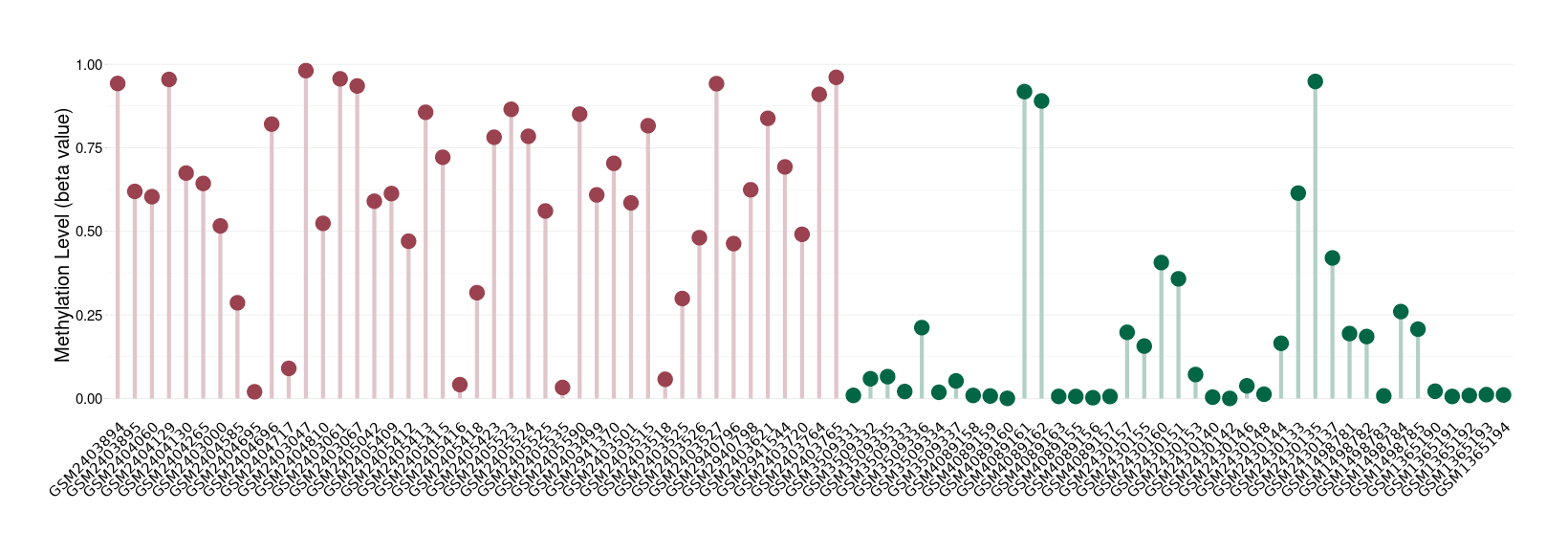 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Brain neuroepithelial tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC34A2 in brain neuroepithelial tumour than that in healthy individual | ||||
Studied Phenotype |
Brain neuroepithelial tumour [ICD-11:2A00.2Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:6.46E-25; Fold-change:0.689345274; Z-score:2.731812707 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
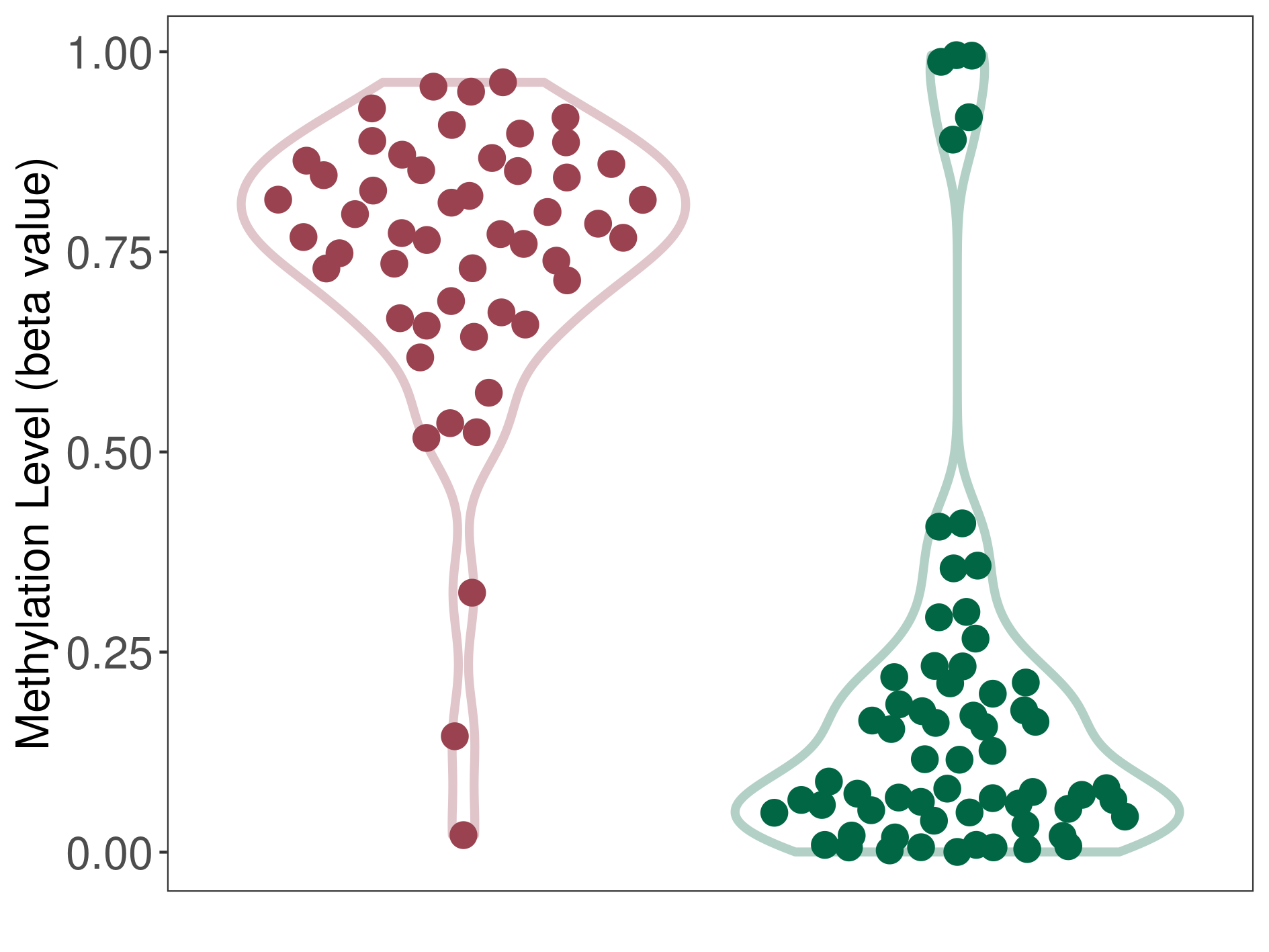
|
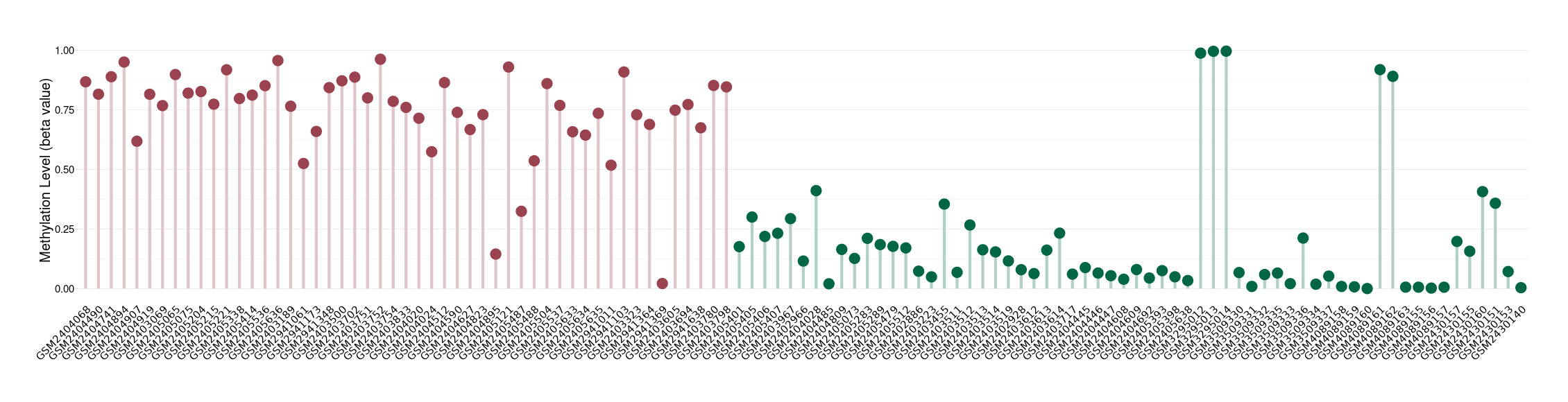 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Diffuse midline glioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC34A2 in diffuse midline glioma than that in healthy individual | ||||
Studied Phenotype |
Diffuse midline glioma [ICD-11:2A00.0Z] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.29E-15; Fold-change:0.352460315; Z-score:1.625528907 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
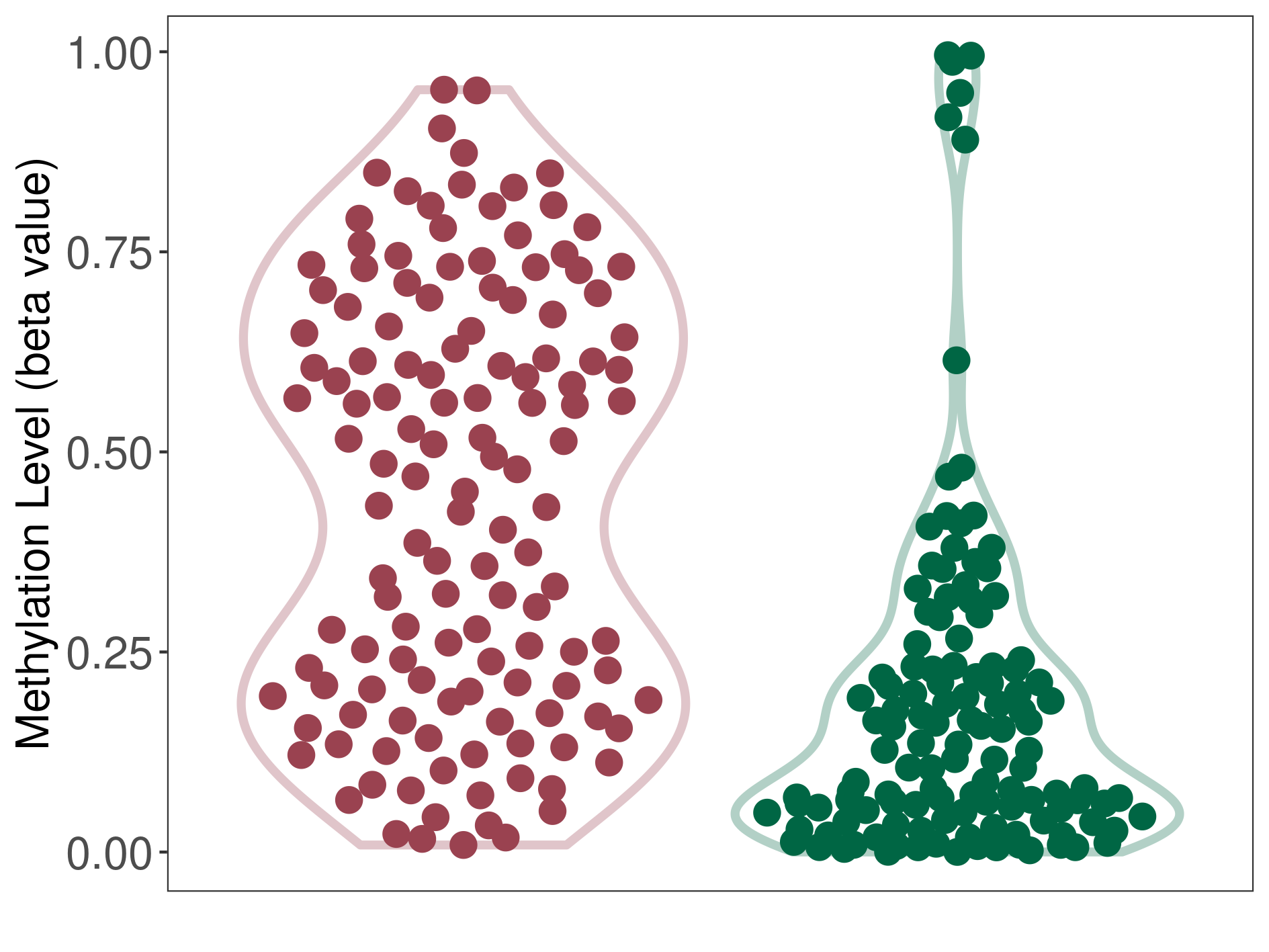
|
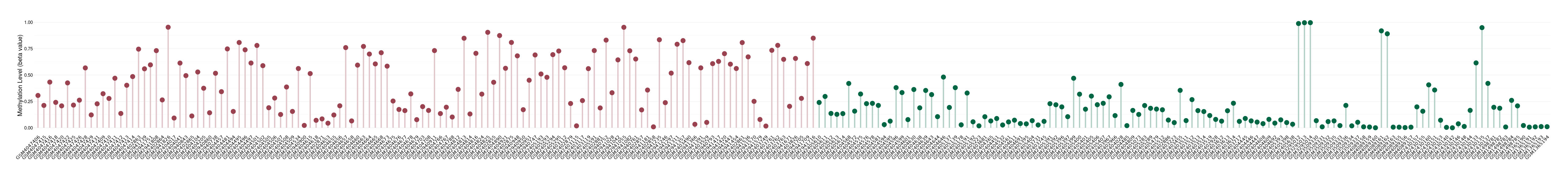 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Glioblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC34A2 in glioblastoma than that in healthy individual | ||||
Studied Phenotype |
Glioblastoma [ICD-11:2A00.00] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:3.55E-05; Fold-change:0.533587211; Z-score:1.473980093 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
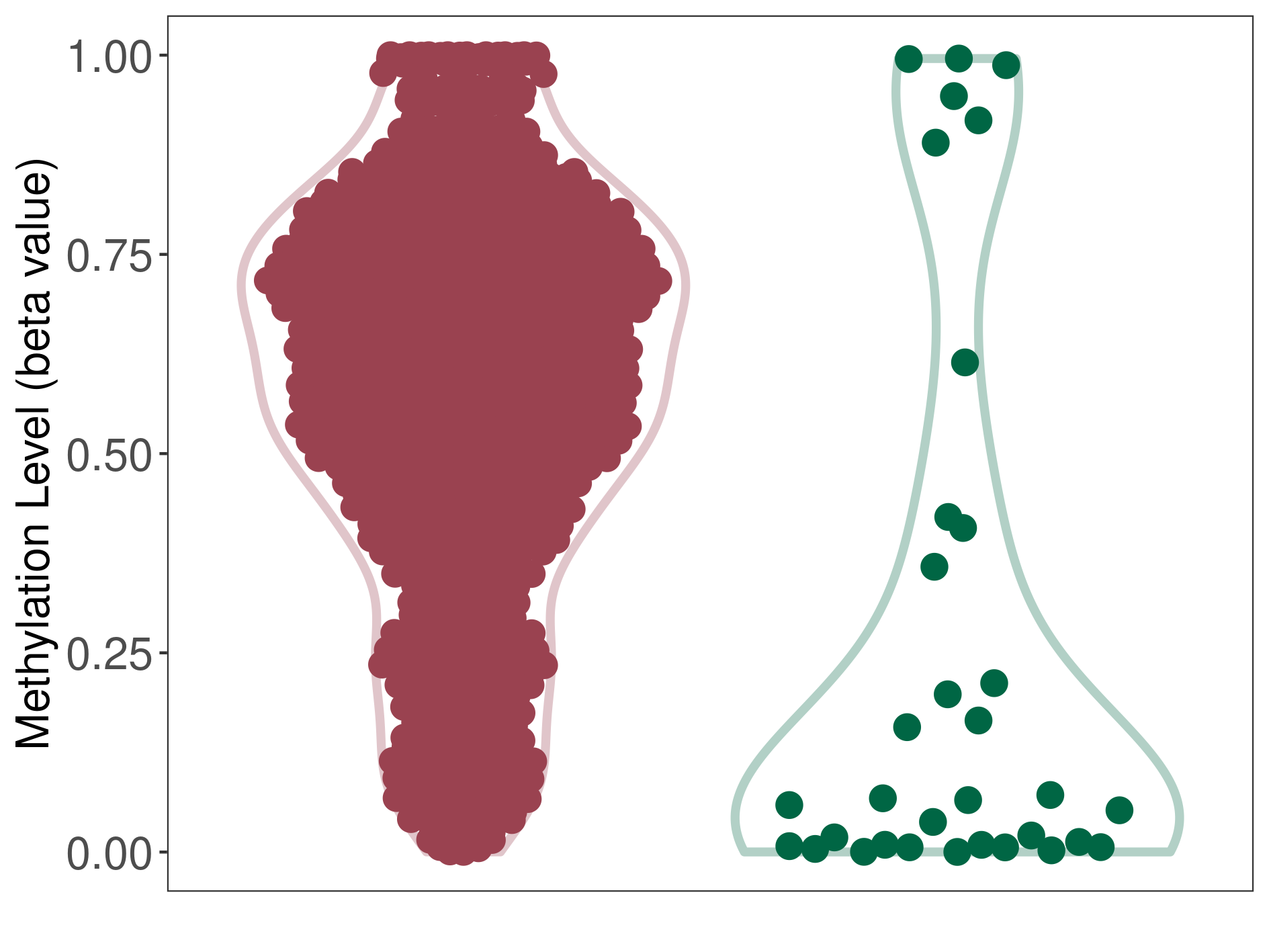
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Lymphoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC34A2 in lymphoma than that in healthy individual | ||||
Studied Phenotype |
Lymphoma [ICD-11:2B30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:5.42E-24; Fold-change:0.643966948; Z-score:2.312055108 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
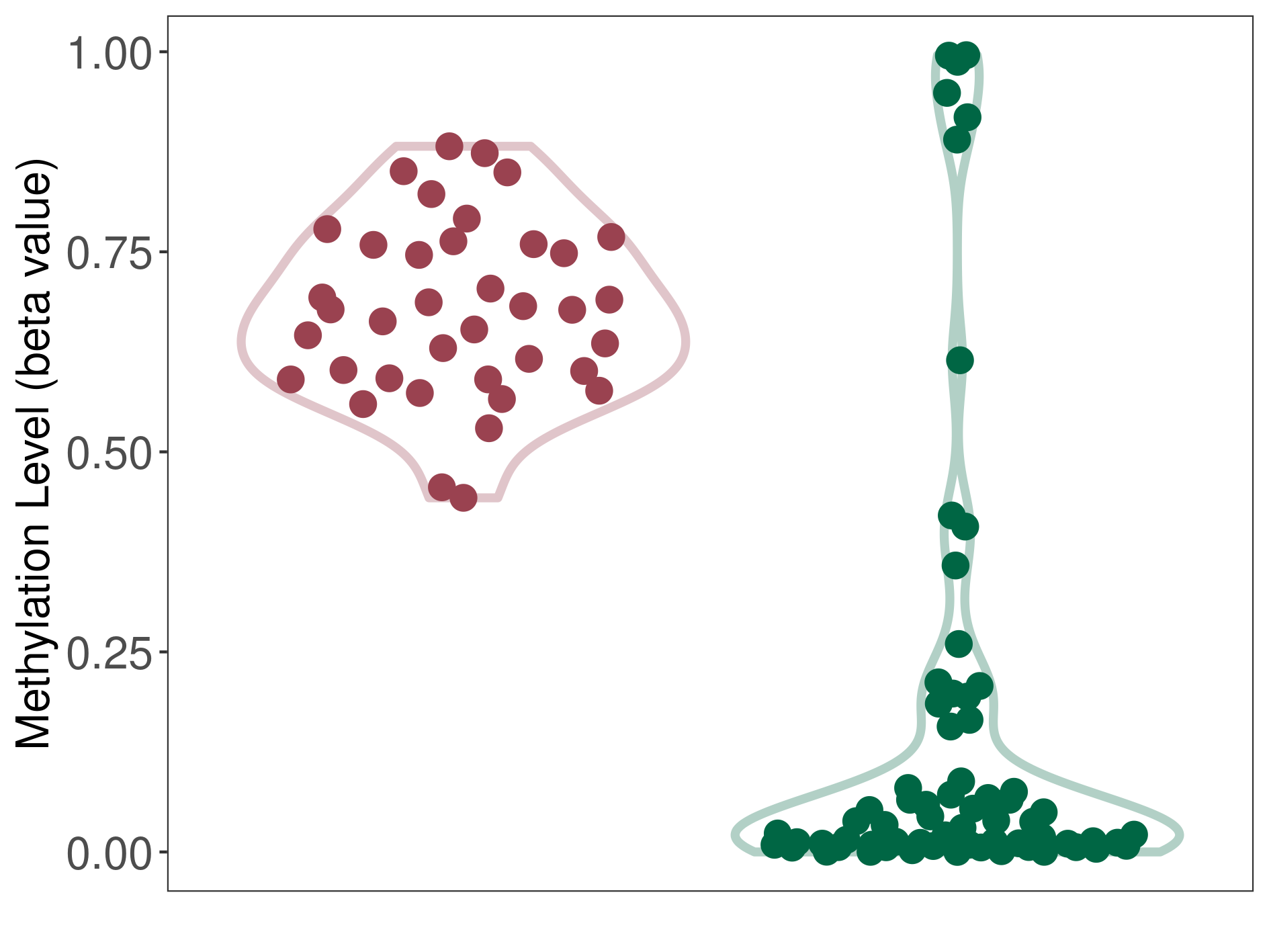
|
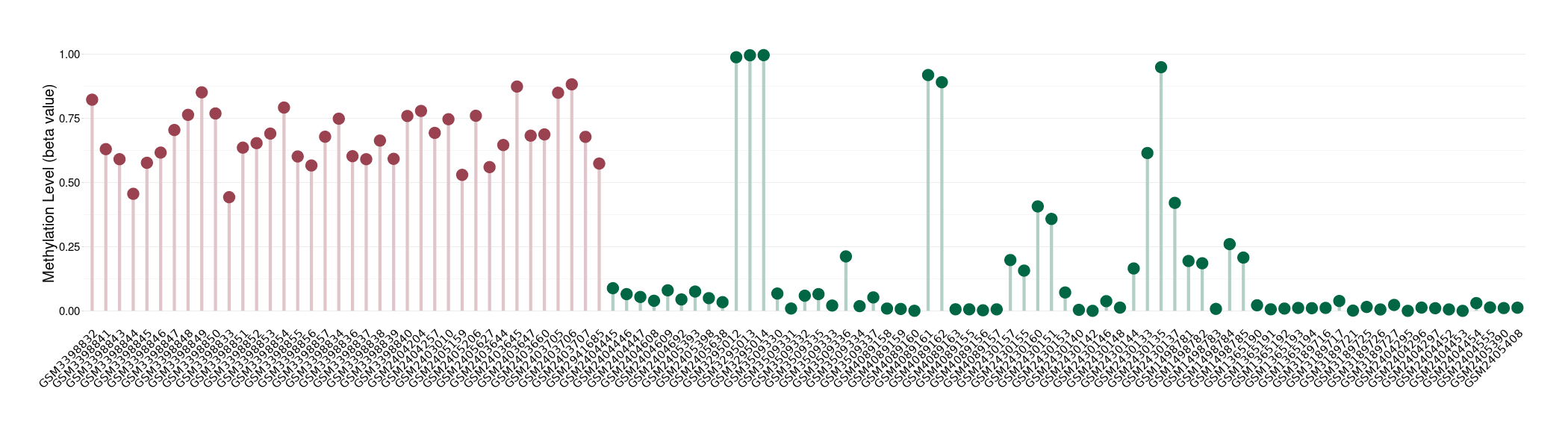 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Malignant astrocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC34A2 in malignant astrocytoma than that in healthy individual | ||||
Studied Phenotype |
Malignant astrocytoma [ICD-11:2A00.12] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:2.06E-46; Fold-change:0.626554291; Z-score:2.760558694 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
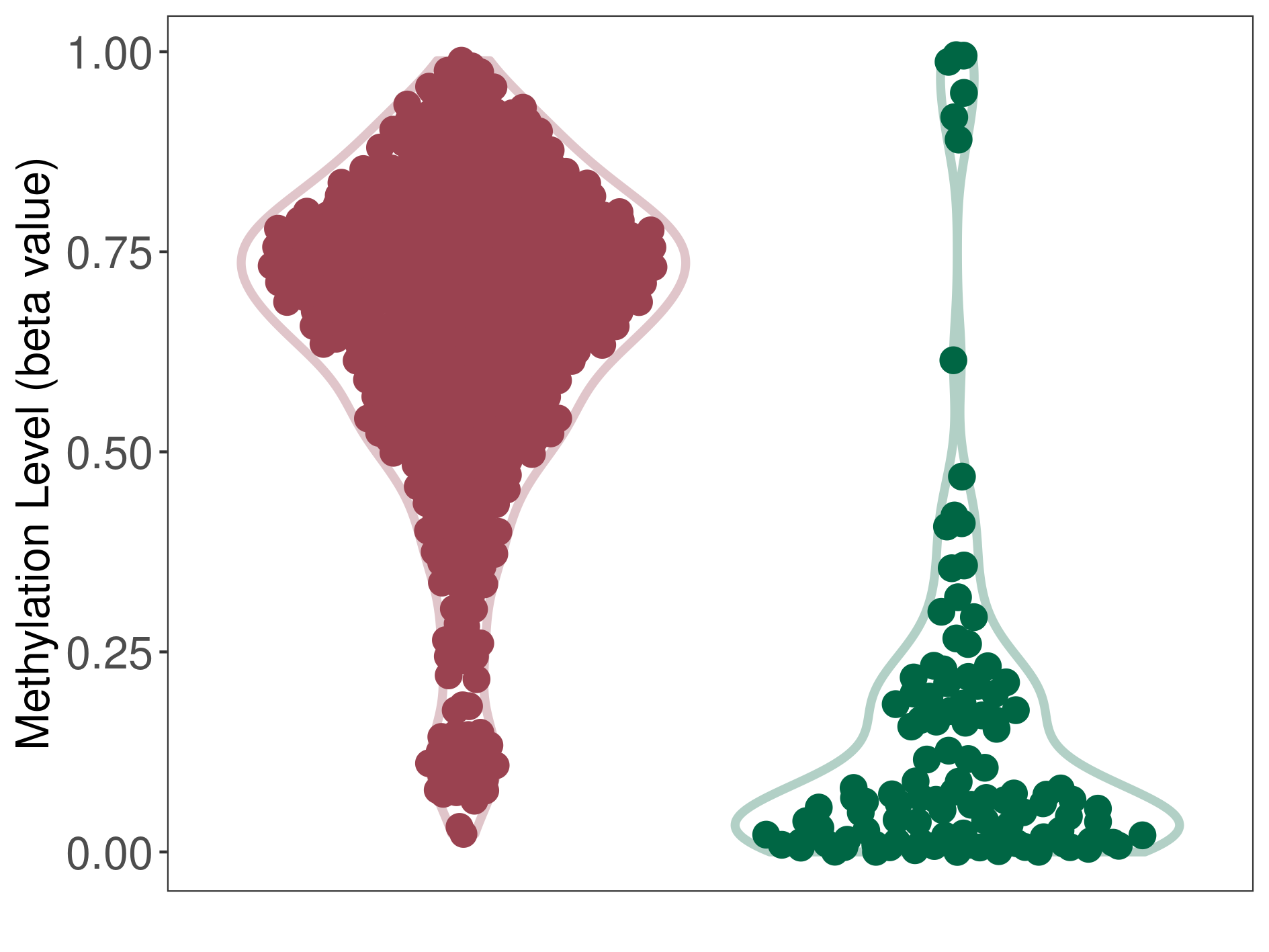
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Multilayered rosettes embryonal tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC34A2 in multilayered rosettes embryonal tumour than that in healthy individual | ||||
Studied Phenotype |
Multilayered rosettes embryonal tumour [ICD-11:2A00.1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:4.58E-21; Fold-change:0.860332864; Z-score:3.293634262 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
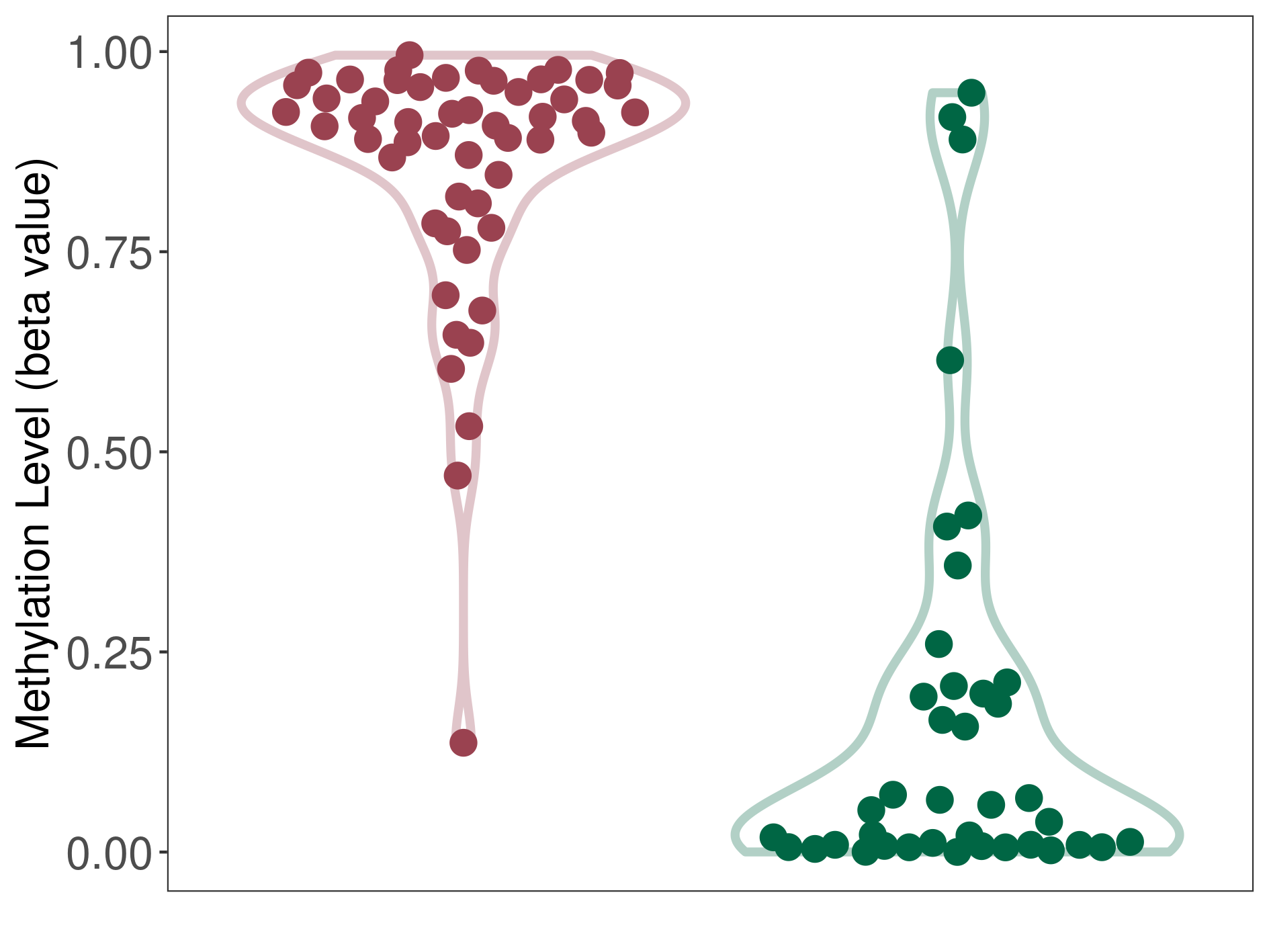
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Oligodendroglial tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC34A2 in oligodendroglial tumour than that in healthy individual | ||||
Studied Phenotype |
Oligodendroglial tumour [ICD-11:2A00.Y1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:6.75E-06; Fold-change:0.483703187; Z-score:5.054048617 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
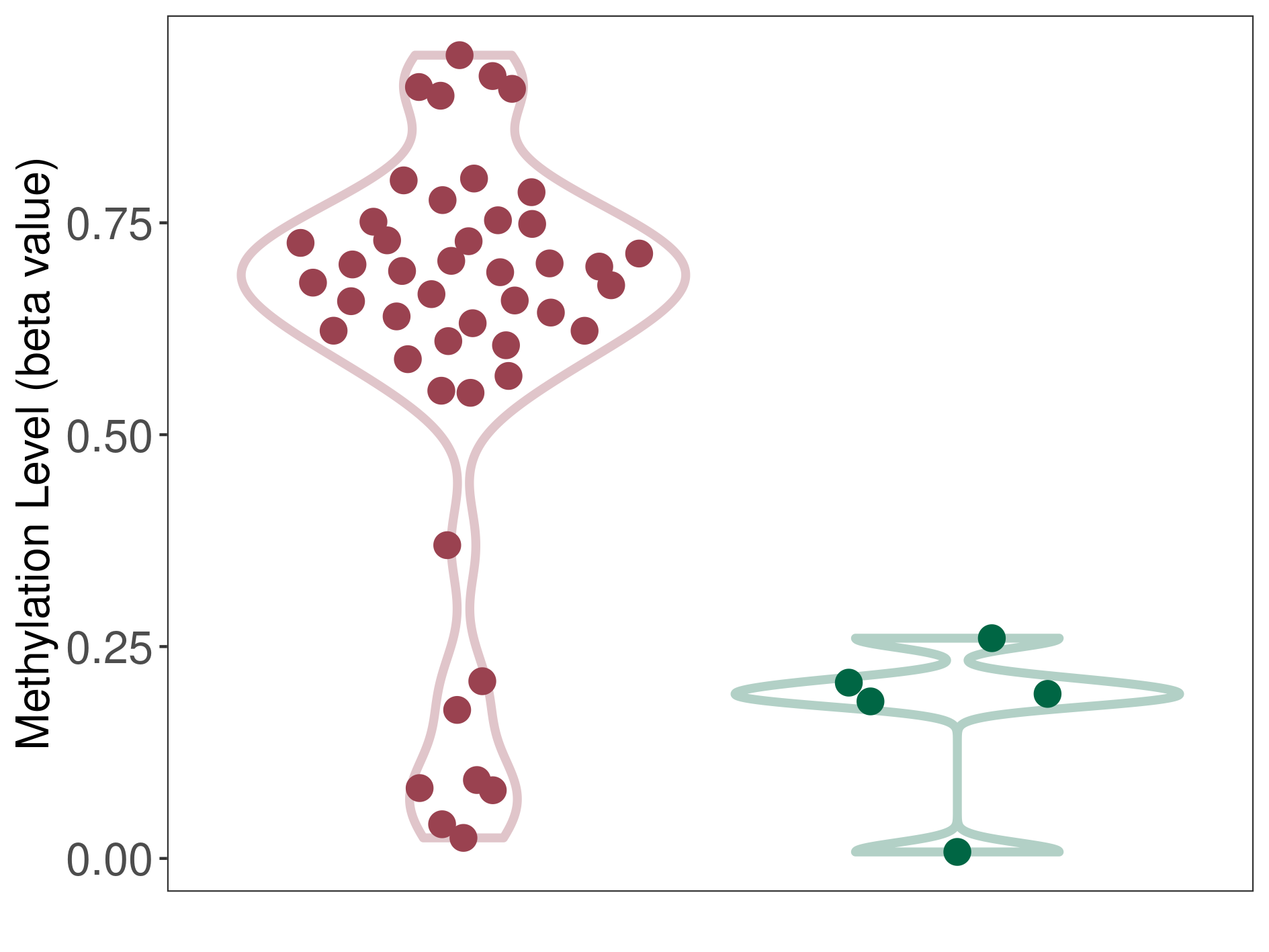
|
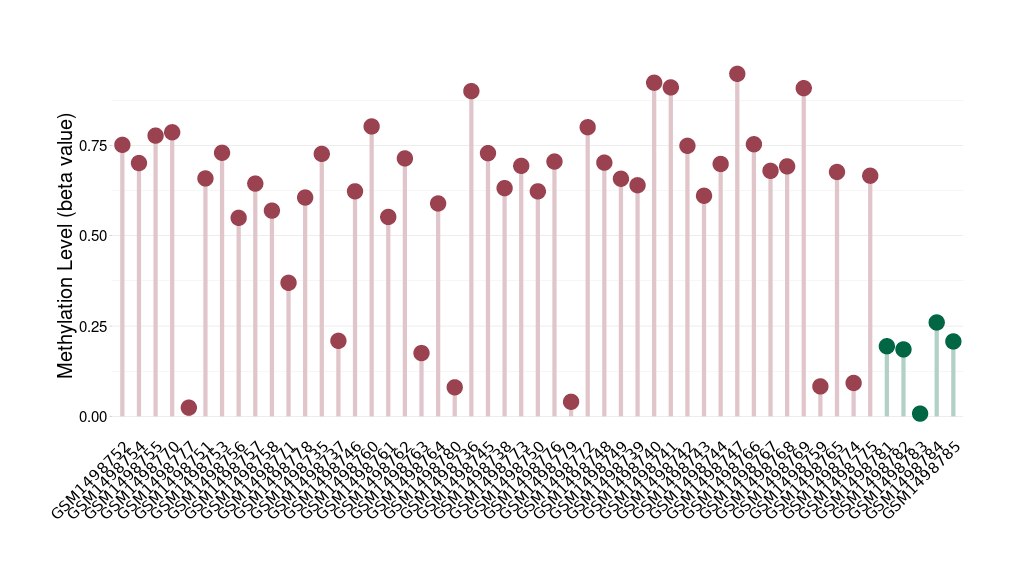 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Oligodendroglioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC34A2 in oligodendroglioma than that in healthy individual | ||||
Studied Phenotype |
Oligodendroglioma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:5.78E-27; Fold-change:0.723744014; Z-score:2.598481258 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
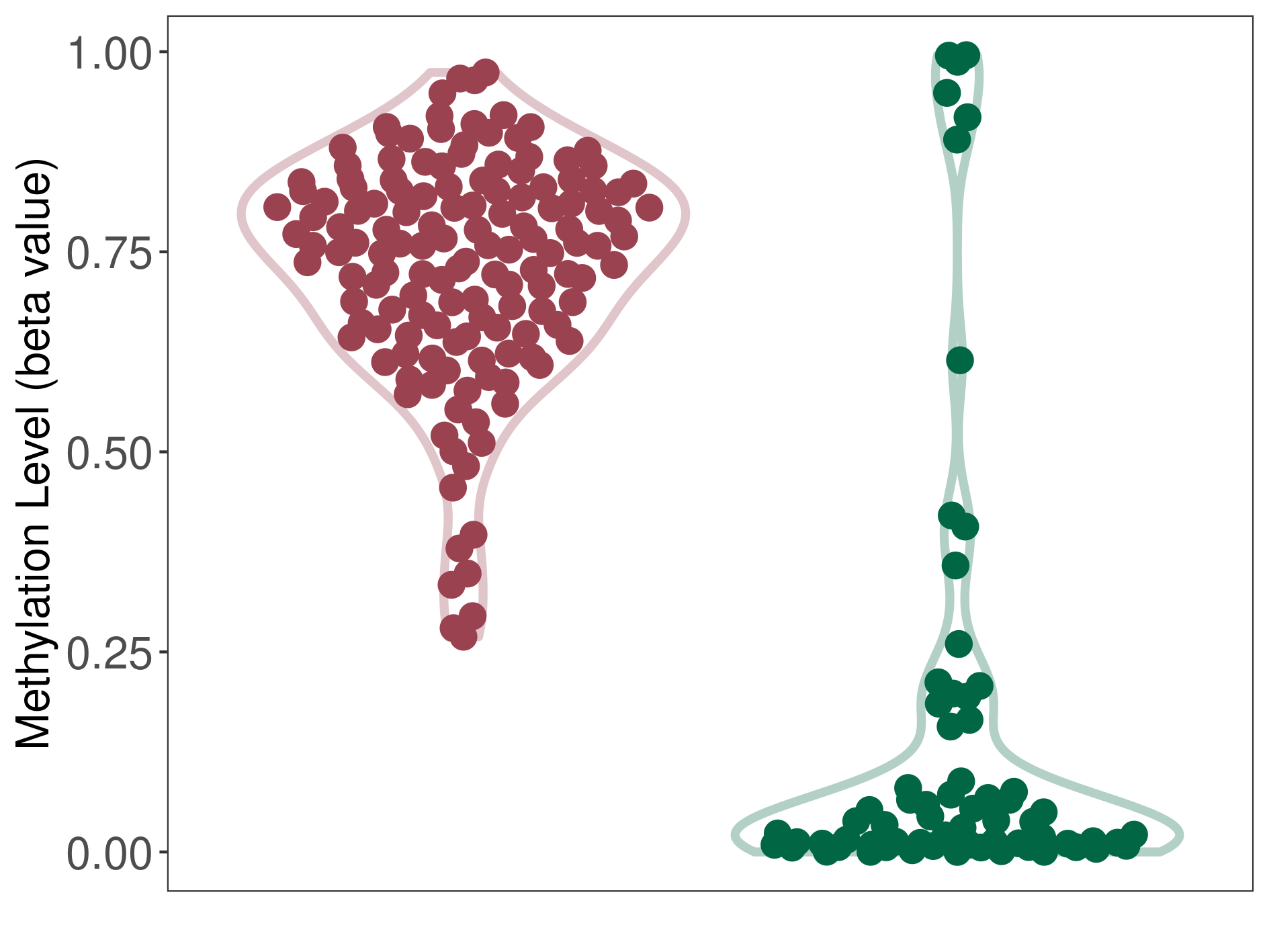
|
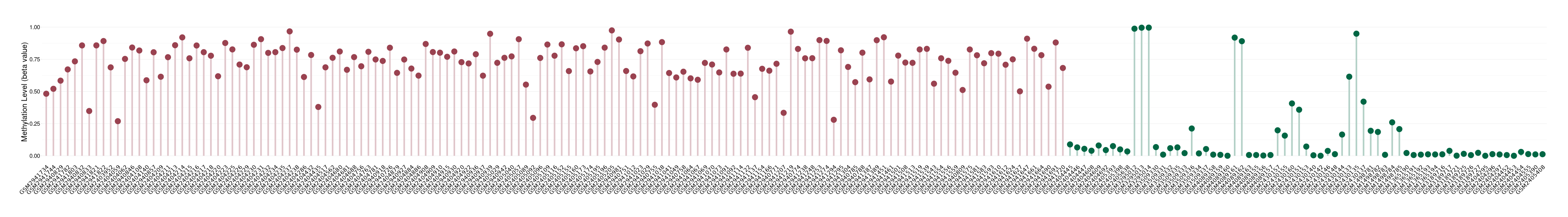 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Posterior fossa ependymoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC34A2 in posterior fossa ependymoma than that in healthy individual | ||||
Studied Phenotype |
Posterior fossa ependymoma [ICD-11:2D50.2] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:5.14E-68; Fold-change:0.51799037; Z-score:2.619124189 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
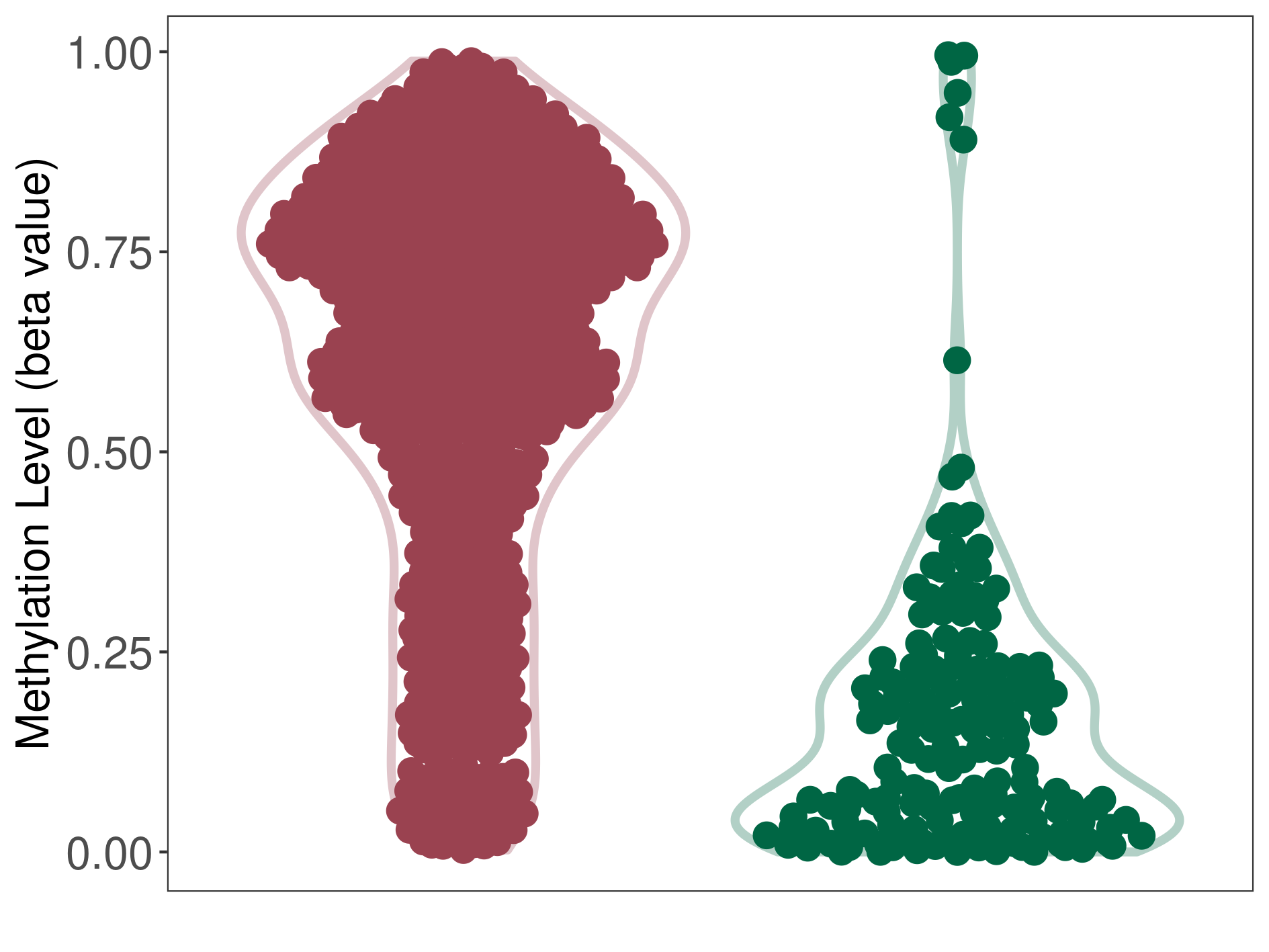
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Prostate cancer metastasis |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC34A2 in prostate cancer metastasis than that in healthy individual | ||||
Studied Phenotype |
Prostate cancer metastasis [ICD-11:2.00E+06] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:7.63E-11; Fold-change:0.66024901; Z-score:52.02292613 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
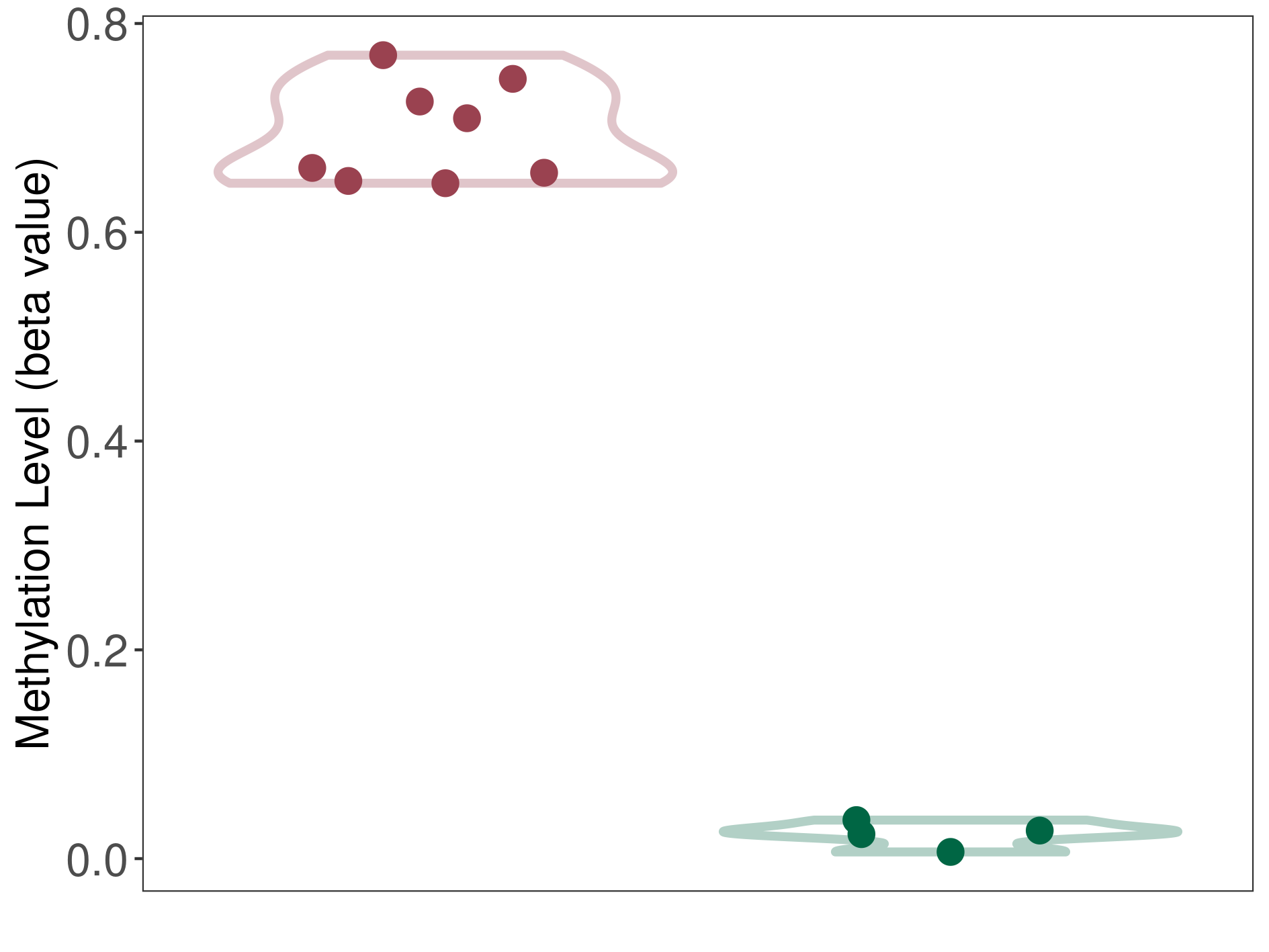
|
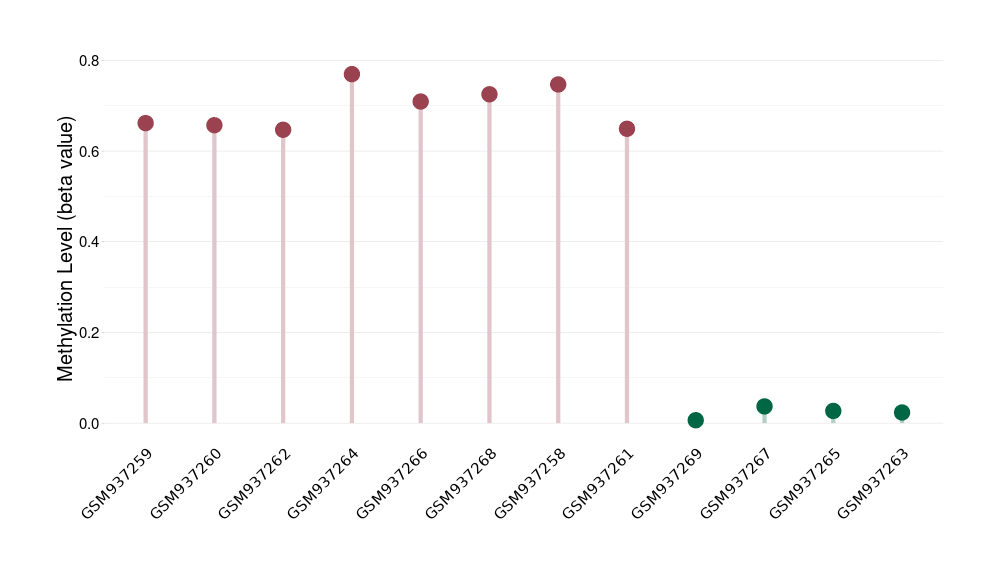 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
RELA YAP fusion ependymoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC34A2 in rela yap fusion ependymoma than that in healthy individual | ||||
Studied Phenotype |
RELA YAP fusion ependymoma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:2.00E-11; Fold-change:0.334348537; Z-score:1.460940436 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
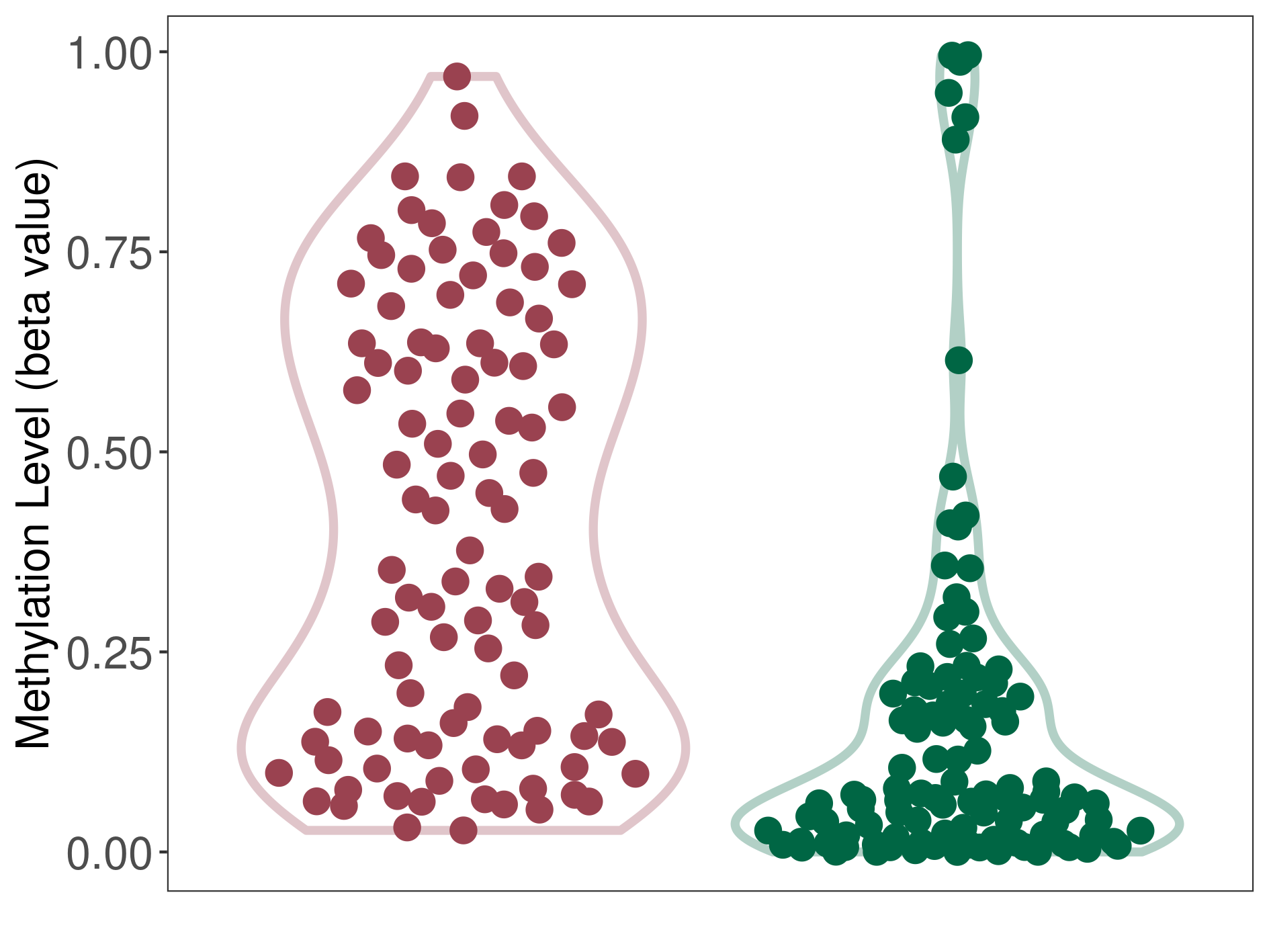
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
microRNA |
|||||
|
Gastric cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Lower expression of miR-939 in gastric cancer (compare with nontumor gastric tissues) | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay | ||
|
Related Molecular Changes | Down regulation ofSLC34A2 | Experiment Method | RT-qPCR | ||
|
miRNA Stemloop ID |
miR-939 | miRNA Mature ID | miR-939-5p | ||
|
miRNA Sequence |
UGGGGAGCUGAGGCUCUGGGGGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Studied Phenotype |
Gastric cancer[ ICD-11:2B72] | ||||
|
Experimental Material |
Patient tissue samples; Multiple cell lines of human | ||||
|
Additional Notes |
miR-939 was down-regulated in gastric cancer and regulated SLC34A2 expression via binding to the SLC34A2 mRNA. | ||||
|
Unclear Phenotype |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
miR-26b directly targets SLC34A2 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-26b | miRNA Mature ID | miR-26b-5p | ||
|
miRNA Sequence |
UUCAAGUAAUUCAGGAUAGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human cervical cancer cell line (Hela) | ||||
|
Epigenetic Phenomenon2 |
miR-335 directly targets SLC34A2 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-335 | miRNA Mature ID | miR-335-5p | ||
|
miRNA Sequence |
UCAAGAGCAAUAACGAAAAAUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon3 |
miR-939 directly targets SLC34A2 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Flow//Immunohistochemistry//Luciferase reporter assay//qRT-PCR//Western blot | ||
|
miRNA Stemloop ID |
miR-939 | miRNA Mature ID | miR-939-5p | ||
|
miRNA Sequence |
UGGGGAGCUGAGGCUCUGGGGGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Patient tissue samples; Multiple cell lines of human | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
