Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0262 Transporter Info | ||||
| Gene Name | SLC2A4 | ||||
| Transporter Name | Glucose transporter type 4, insulin-responsive | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Depression |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Hypermethylation of SLC2A4 in depressive disorder | [ 1 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Bisulfite sequencing | ||
|
Related Molecular Changes | Down regulation ofSLC2A4 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Additional Notes |
The methylation level of the GLUT4 significantly increased in depressed inpatients compared to healthy comparison subjects. | ||||
|
Colon cancer |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC2A4 in colon adenocarcinoma | [ 2 ] | |||
|
Location |
5'UTR (cg12718793) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.83E+00 | Statistic Test | p-value:1.18E-03; Z-score:-2.13E+00 | ||
|
Methylation in Case |
1.52E-01 (Median) | Methylation in Control | 2.79E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC2A4 in colon adenocarcinoma | [ 2 ] | |||
|
Location |
1stExon (cg17793621) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.59E+00 | Statistic Test | p-value:1.32E-06; Z-score:2.16E+00 | ||
|
Methylation in Case |
5.24E-01 (Median) | Methylation in Control | 3.29E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC2A4 in colon adenocarcinoma | [ 2 ] | |||
|
Location |
1stExon (cg15012572) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.32E+00 | Statistic Test | p-value:4.73E-03; Z-score:1.18E+00 | ||
|
Methylation in Case |
3.87E-02 (Median) | Methylation in Control | 2.94E-02 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC2A4 in colon adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg19930802) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:7.54E-04; Z-score:-2.16E+00 | ||
|
Methylation in Case |
8.70E-01 (Median) | Methylation in Control | 9.05E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC2A4 in bladder cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg02694091) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:2.83E-02; Z-score:-1.55E+00 | ||
|
Methylation in Case |
6.97E-02 (Median) | Methylation in Control | 7.79E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC2A4 in bladder cancer | [ 3 ] | |||
|
Location |
TSS200 (cg09103232) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.27E+00 | Statistic Test | p-value:1.62E-02; Z-score:-1.58E+00 | ||
|
Methylation in Case |
6.65E-02 (Median) | Methylation in Control | 8.43E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC2A4 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg06891043) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.31E+00 | Statistic Test | p-value:1.06E-04; Z-score:-3.95E+00 | ||
|
Methylation in Case |
4.65E-01 (Median) | Methylation in Control | 6.10E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC2A4 in bladder cancer | [ 3 ] | |||
|
Location |
3'UTR (cg03670302) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.55E+00 | Statistic Test | p-value:7.39E-11; Z-score:-9.71E+00 | ||
|
Methylation in Case |
5.74E-01 (Median) | Methylation in Control | 8.91E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC2A4 in breast cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg02694091) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:7.72E-03; Z-score:-5.94E-01 | ||
|
Methylation in Case |
5.95E-02 (Median) | Methylation in Control | 7.01E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC2A4 in breast cancer | [ 4 ] | |||
|
Location |
TSS200 (cg07287120) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:3.58E-02; Z-score:-2.67E-01 | ||
|
Methylation in Case |
5.48E-02 (Median) | Methylation in Control | 6.04E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC2A4 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg27067158) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.28E+00 | Statistic Test | p-value:2.01E-23; Z-score:3.87E+00 | ||
|
Methylation in Case |
7.60E-01 (Median) | Methylation in Control | 5.96E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC2A4 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg21994579) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.16E+00 | Statistic Test | p-value:8.82E-05; Z-score:6.70E-01 | ||
|
Methylation in Case |
1.10E-01 (Median) | Methylation in Control | 9.48E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC2A4 in clear cell renal cell carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg00064950) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:3.27E-02; Z-score:4.49E-01 | ||
|
Methylation in Case |
3.76E-02 (Median) | Methylation in Control | 3.45E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC2A4 in clear cell renal cell carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg07723558) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:4.14E-02; Z-score:1.79E-01 | ||
|
Methylation in Case |
4.34E-02 (Median) | Methylation in Control | 4.22E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC2A4 in clear cell renal cell carcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg09103232) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.41E+00 | Statistic Test | p-value:4.70E-03; Z-score:-1.11E+00 | ||
|
Methylation in Case |
2.07E-02 (Median) | Methylation in Control | 2.93E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC2A4 in clear cell renal cell carcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg13787272) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:7.77E-03; Z-score:-9.60E-01 | ||
|
Methylation in Case |
2.24E-02 (Median) | Methylation in Control | 3.06E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC2A4 in clear cell renal cell carcinoma | [ 5 ] | |||
|
Location |
Body (cg06891043) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:1.00E-06; Z-score:-2.21E+00 | ||
|
Methylation in Case |
7.93E-01 (Median) | Methylation in Control | 8.57E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC2A4 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS1500 (cg07723558) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:3.10E-02; Z-score:5.21E-01 | ||
|
Methylation in Case |
1.23E-01 (Median) | Methylation in Control | 1.15E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC2A4 in colorectal cancer | [ 6 ] | |||
|
Location |
TSS200 (cg25362068) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.20E+00 | Statistic Test | p-value:3.05E-02; Z-score:-3.61E-01 | ||
|
Methylation in Case |
2.26E-02 (Median) | Methylation in Control | 2.70E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC2A4 in colorectal cancer | [ 6 ] | |||
|
Location |
Body (cg21994579) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:9.88E-06; Z-score:9.32E-01 | ||
|
Methylation in Case |
1.78E-01 (Median) | Methylation in Control | 1.48E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Pancretic ductal adenocarcinoma |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC2A4 in pancretic ductal adenocarcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg15971010) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.23E+00 | Statistic Test | p-value:5.90E-07; Z-score:1.39E+00 | ||
|
Methylation in Case |
3.82E-01 (Median) | Methylation in Control | 3.11E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC2A4 in pancretic ductal adenocarcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg06463759) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:1.53E-02; Z-score:-8.26E-01 | ||
|
Methylation in Case |
6.70E-02 (Median) | Methylation in Control | 9.14E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC2A4 in pancretic ductal adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg22574986) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:1.48E-03; Z-score:5.74E-01 | ||
|
Methylation in Case |
8.68E-02 (Median) | Methylation in Control | 7.40E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC2A4 in prostate cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg07903677) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.51E+00 | Statistic Test | p-value:2.72E-03; Z-score:6.45E+00 | ||
|
Methylation in Case |
6.99E-01 (Median) | Methylation in Control | 4.64E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC2A4 in prostate cancer | [ 8 ] | |||
|
Location |
Body (cg15266604) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:7.79E-04; Z-score:4.13E+00 | ||
|
Methylation in Case |
9.48E-01 (Median) | Methylation in Control | 9.08E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Systemic lupus erythematosus |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC2A4 in systemic lupus erythematosus | [ 9 ] | |||
|
Location |
TSS1500 (cg00064950) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.47E-02; Z-score:-1.36E-01 | ||
|
Methylation in Case |
9.53E-02 (Median) | Methylation in Control | 9.76E-02 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC2A4 in systemic lupus erythematosus | [ 9 ] | |||
|
Location |
TSS1500 (cg02694091) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:2.62E-02; Z-score:-1.19E-01 | ||
|
Methylation in Case |
7.69E-02 (Median) | Methylation in Control | 7.87E-02 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC2A4 in systemic lupus erythematosus | [ 9 ] | |||
|
Location |
TSS200 (cg13787272) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.99E-04; Z-score:-1.15E-01 | ||
|
Methylation in Case |
8.43E-02 (Median) | Methylation in Control | 8.61E-02 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC2A4 in systemic lupus erythematosus | [ 9 ] | |||
|
Location |
Body (cg06891043) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:4.37E-02; Z-score:-2.79E-01 | ||
|
Methylation in Case |
7.05E-01 (Median) | Methylation in Control | 7.23E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC2A4 in panic disorder | [ 10 ] | |||
|
Location |
TSS200 (cg07287120) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:9.93E-01 | Statistic Test | p-value:3.22E-02; Z-score:1.48E-01 | ||
|
Methylation in Case |
-4.98E+00 (Median) | Methylation in Control | -5.01E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC2A4 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
TSS200 (cg26480745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:2.47E-03; Z-score:-5.32E-01 | ||
|
Methylation in Case |
1.45E-01 (Median) | Methylation in Control | 1.60E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Atypical teratoid rhabdoid tumor |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC2A4 in atypical teratoid rhabdoid tumor | [ 12 ] | |||
|
Location |
Body (cg06891043) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:3.08E-03; Z-score:7.14E-01 | ||
|
Methylation in Case |
7.98E-01 (Median) | Methylation in Control | 7.27E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC2A4 in atypical teratoid rhabdoid tumor | [ 12 ] | |||
|
Location |
3'UTR (cg03670302) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.26E+00 | Statistic Test | p-value:1.31E-14; Z-score:-2.60E+00 | ||
|
Methylation in Case |
6.85E-01 (Median) | Methylation in Control | 8.60E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC2A4 in hepatocellular carcinoma | [ 13 ] | |||
|
Location |
Body (cg03924551) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.43E+00 | Statistic Test | p-value:1.47E-15; Z-score:-4.73E+00 | ||
|
Methylation in Case |
5.42E-01 (Median) | Methylation in Control | 7.77E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC2A4 in hepatocellular carcinoma | [ 13 ] | |||
|
Location |
Body (cg13532410) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.47E+00 | Statistic Test | p-value:1.13E-12; Z-score:-1.76E+00 | ||
|
Methylation in Case |
3.34E-01 (Median) | Methylation in Control | 4.91E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC2A4 in hepatocellular carcinoma | [ 13 ] | |||
|
Location |
Body (cg27067158) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:6.68E-03; Z-score:-7.42E-01 | ||
|
Methylation in Case |
8.27E-01 (Median) | Methylation in Control | 8.38E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC2A4 in hepatocellular carcinoma | [ 13 ] | |||
|
Location |
3'UTR (cg03670302) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.22E-06; Z-score:-1.79E+00 | ||
|
Methylation in Case |
8.91E-01 (Median) | Methylation in Control | 9.28E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
HIV infection |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC2A4 in HIV infection | [ 14 ] | |||
|
Location |
Body (cg21994579) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.46E+00 | Statistic Test | p-value:6.25E-05; Z-score:1.76E+00 | ||
|
Methylation in Case |
1.27E-01 (Median) | Methylation in Control | 8.71E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC2A4 in HIV infection | [ 14 ] | |||
|
Location |
3'UTR (cg03670302) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:3.09E-13; Z-score:2.13E+00 | ||
|
Methylation in Case |
9.44E-01 (Median) | Methylation in Control | 9.00E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC2A4 in lung adenocarcinoma | [ 15 ] | |||
|
Location |
Body (cg06891043) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:2.16E-02; Z-score:-1.23E+00 | ||
|
Methylation in Case |
7.14E-01 (Median) | Methylation in Control | 7.54E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC2A4 in lung adenocarcinoma | [ 15 ] | |||
|
Location |
3'UTR (cg03670302) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:8.98E-03; Z-score:-1.79E+00 | ||
|
Methylation in Case |
8.86E-01 (Median) | Methylation in Control | 9.09E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lymphoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC2A4 in lymphoma than that in healthy individual | ||||
Studied Phenotype |
Lymphoma [ICD-11:2B30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.30E-11; Fold-change:0.496755918; Z-score:25.16026682 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
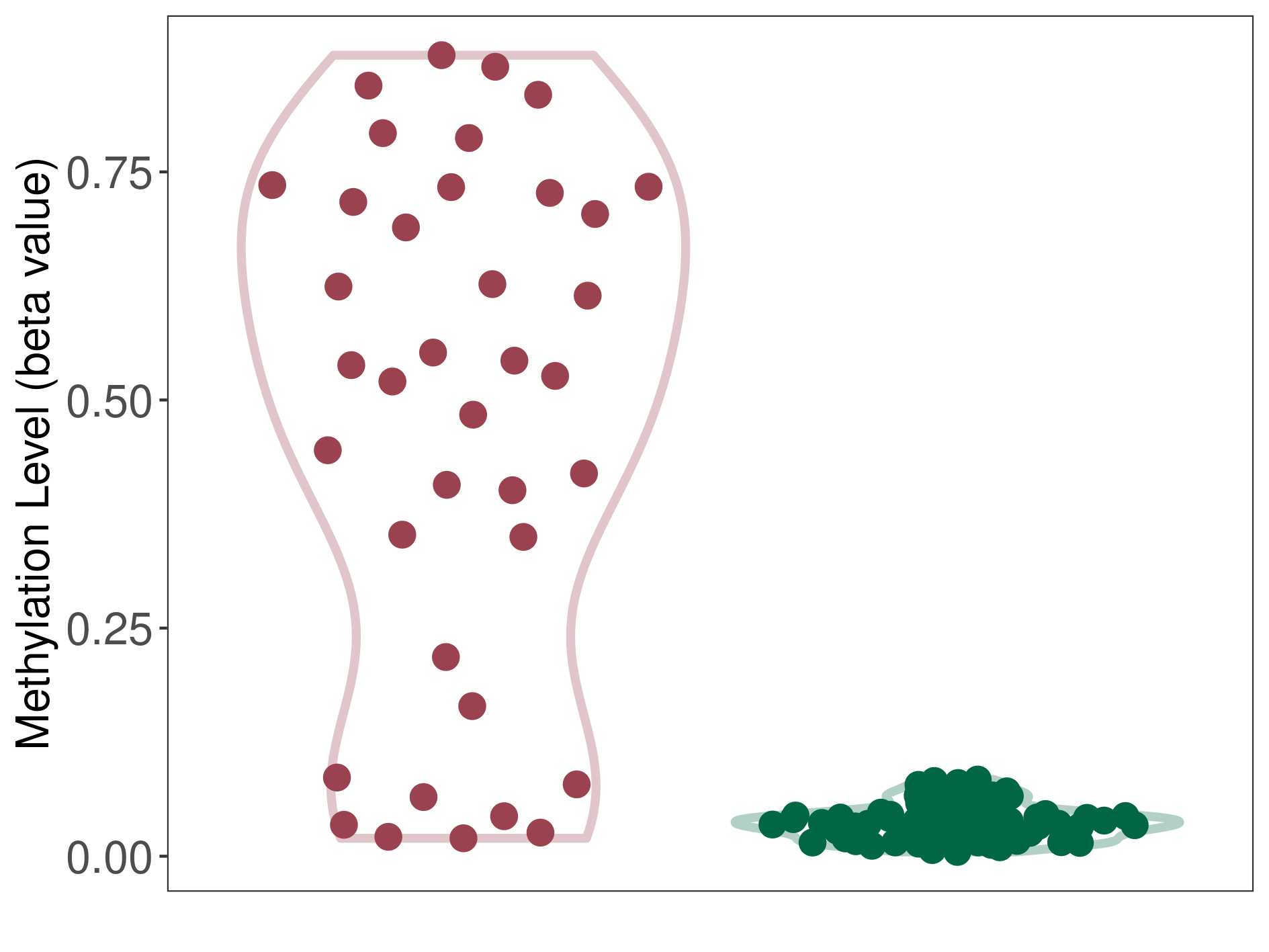
|
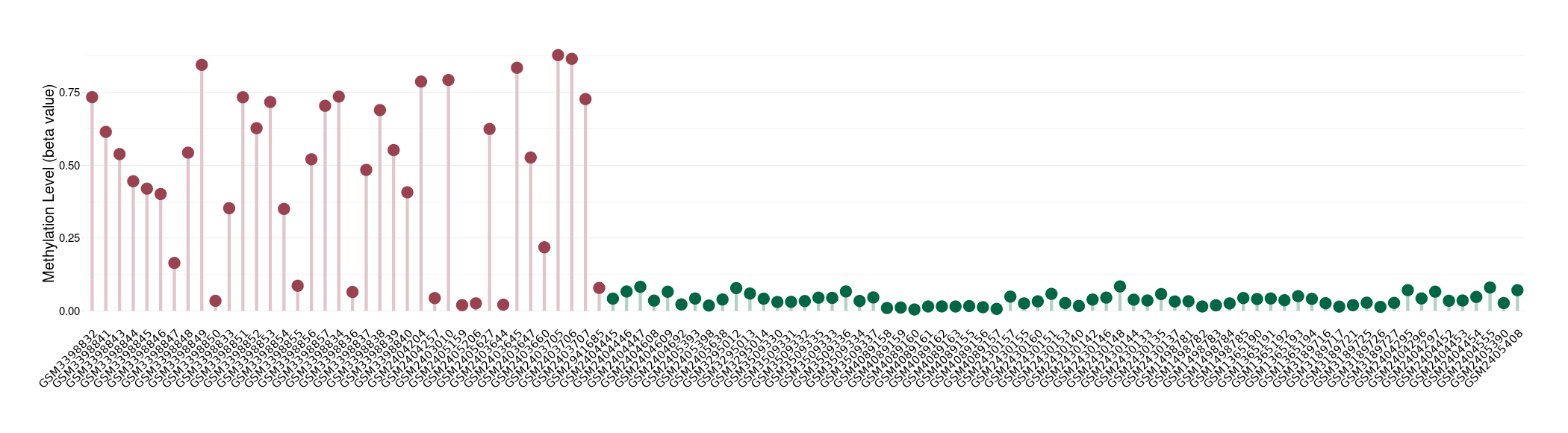 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Histone acetylation |
|||||
|
Non-small cell lung cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Hypoacetylation of SLC2A4 in non-small cell lung cancer (compare with Vorinostat-treatment counterpart cells) | [ 16 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes | Down regulation ofSLC2A4 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Non-small cell lung cancer[ ICD-11:2C25.4] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
SLC2A4 expression can be induced or increased following inhibition of histone deacetylases in non-small cell lung cancer cell lines. | ||||
|
Type 1 diabetes |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Hypoacetylation of SLC2A4 in type 1 diabete (compare with counterpart normal rat cells) | [ 17 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes | Down regulation ofSLC2A4 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Type 1 diabetes[ ICD-11:5A10] | ||||
|
Experimental Material |
Model organism in vivo (mouse) | ||||
|
Additional Notes |
The Type 1 diabetes reduced the H3Kac which would decrease the binding of transcription factors. | ||||
|
Type 2 diabetes |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Hypoacetylation of SLC2A4 in type 2 diabete (compare with counterpart normal rat cells) | [ 17 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes | Down regulation ofSLC2A4 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Type 2 diabetes[ ICD-11:5A11] | ||||
|
Experimental Material |
Model organism in vivo (mouse) | ||||
|
Additional Notes |
The Type 2 diabetes reduced the H3Kac which would decrease the binding of transcription factors. | ||||
|
Histone trimethylation |
|||||
|
Type 1 diabetes |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Higher level of histone trimethylation of SLC2A4 in type 1 diabete (compare with counterpart normal rat cells) | [ 17 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone trimethylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes | Down regulation ofSLC2A4 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Type 1 diabetes[ ICD-11:5A10] | ||||
|
Experimental Material |
Model organism in vivo (mouse) | ||||
|
Additional Notes |
The Type 1 diabetes reduced the H3Kac and increased the H3K9me3 content, and insulin treatment reversed only the H3K9me3 modification. | ||||
|
Type 2 diabetes |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Lower level of histone trimethylation of SLC2A4 in type 2 diabete (compare with counterpart normal rat cells) | [ 17 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone trimethylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes | Down regulation ofSLC2A4 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Type 2 diabetes[ ICD-11:5A11] | ||||
|
Experimental Material |
Model organism in vivo (mouse) | ||||
|
Additional Notes |
The Type 2 diabetes reduced the H3Kac and increased the H3K9me3 content, and insulin treatment reversed only the H3K9me3 modification. | ||||
|
microRNA |
|||||
|
Unclear Phenotype |
28 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
miR-1286 directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1286 | miRNA Mature ID | miR-1286 | ||
|
miRNA Sequence |
UGCAGGACCAAGAUGAGCCCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon2 |
miR-1321 directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1321 | miRNA Mature ID | miR-1321 | ||
|
miRNA Sequence |
CAGGGAGGUGAAUGUGAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon3 |
miR-3121 directly targets SLC2A4 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3121 | miRNA Mature ID | miR-3121-5p | ||
|
miRNA Sequence |
UCCUUUGCCUAUUCUAUUUAAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon4 |
miR-335 directly targets SLC2A4 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-335 | miRNA Mature ID | miR-335-5p | ||
|
miRNA Sequence |
UCAAGAGCAAUAACGAAAAAUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon5 |
miR-4436a directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4436a | miRNA Mature ID | miR-4436a | ||
|
miRNA Sequence |
GCAGGACAGGCAGAAGUGGAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon6 |
miR-4510 directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4510 | miRNA Mature ID | miR-4510 | ||
|
miRNA Sequence |
UGAGGGAGUAGGAUGUAUGGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon7 |
miR-4709 directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4709 | miRNA Mature ID | miR-4709-3p | ||
|
miRNA Sequence |
UUGAAGAGGAGGUGCUCUGUAGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon8 |
miR-4739 directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4739 | miRNA Mature ID | miR-4739 | ||
|
miRNA Sequence |
AAGGGAGGAGGAGCGGAGGGGCCCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon9 |
miR-4756 directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4756 | miRNA Mature ID | miR-4756-5p | ||
|
miRNA Sequence |
CAGGGAGGCGCUCACUCUCUGCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon10 |
miR-5000 directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-5000 | miRNA Mature ID | miR-5000-3p | ||
|
miRNA Sequence |
UCAGGACACUUCUGAACUUGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon11 |
miR-548ac directly targets SLC2A4 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-548ac | miRNA Mature ID | miR-548ac | ||
|
miRNA Sequence |
CAAAAACCGGCAAUUACUUUUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon12 |
miR-548as directly targets SLC2A4 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-548as | miRNA Mature ID | miR-548as-3p | ||
|
miRNA Sequence |
UAAAACCCACAAUUAUGUUUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon13 |
miR-548bb directly targets SLC2A4 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-548bb | miRNA Mature ID | miR-548bb-3p | ||
|
miRNA Sequence |
CAAAAACCAUAGUUACUUUUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon14 |
miR-548d directly targets SLC2A4 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-548d | miRNA Mature ID | miR-548d-3p | ||
|
miRNA Sequence |
CAAAAACCACAGUUUCUUUUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon15 |
miR-548h directly targets SLC2A4 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-548h | miRNA Mature ID | miR-548h-3p | ||
|
miRNA Sequence |
CAAAAACCGCAAUUACUUUUGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon16 |
miR-548z directly targets SLC2A4 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-548z | miRNA Mature ID | miR-548z | ||
|
miRNA Sequence |
CAAAAACCGCAAUUACUUUUGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon17 |
miR-6077 directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6077 | miRNA Mature ID | miR-6077 | ||
|
miRNA Sequence |
GGGAAGAGCUGUACGGCCUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon18 |
miR-6127 directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6127 | miRNA Mature ID | miR-6127 | ||
|
miRNA Sequence |
UGAGGGAGUGGGUGGGAGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon19 |
miR-6129 directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6129 | miRNA Mature ID | miR-6129 | ||
|
miRNA Sequence |
UGAGGGAGUUGGGUGUAUA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon20 |
miR-6130 directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6130 | miRNA Mature ID | miR-6130 | ||
|
miRNA Sequence |
UGAGGGAGUGGAUUGUAUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon21 |
miR-6133 directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6133 | miRNA Mature ID | miR-6133 | ||
|
miRNA Sequence |
UGAGGGAGGAGGUUGGGUA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon22 |
miR-6771 directly targets SLC2A4 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6771 | miRNA Mature ID | miR-6771-3p | ||
|
miRNA Sequence |
CAAACCCCUGUCUACCCGCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Left ventricular cardiac tissues of human | ||||
|
Epigenetic Phenomenon23 |
miR-6782 directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6782 | miRNA Mature ID | miR-6782-5p | ||
|
miRNA Sequence |
UAGGGGUGGGGGAAUUCAGGGGUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon24 |
miR-6873 directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6873 | miRNA Mature ID | miR-6873-5p | ||
|
miRNA Sequence |
CAGAGGGAAUACAGAGGGCAAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon25 |
miR-6880 directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6880 | miRNA Mature ID | miR-6880-5p | ||
|
miRNA Sequence |
UGGUGGAGGAAGAGGGCAGCUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon26 |
miR-7106 directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-7106 | miRNA Mature ID | miR-7106-5p | ||
|
miRNA Sequence |
UGGGAGGAGGGGAUCUUGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon27 |
miR-766 directly targets SLC2A4 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-766 | miRNA Mature ID | miR-766-5p | ||
|
miRNA Sequence |
AGGAGGAAUUGGUGCUGGUCUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon28 |
miR-92a-1 directly targets SLC2A4 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | CLASH | ||
|
miRNA Stemloop ID |
miR-92a-1 | miRNA Mature ID | miR-92a-1-5p | ||
|
miRNA Sequence |
AGGUUGGGAUCGGUUGCAAUGCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
