Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0143 Transporter Info | ||||
| Gene Name | SLC22A17 | ||||
| Transporter Name | Brain-type organic cation transporter | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Bladder cancer |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC22A17 in bladder cancer | [ 1 ] | |||
|
Location |
TSS1500 (cg26922038) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.27E+00 | Statistic Test | p-value:8.66E-13; Z-score:-1.40E+01 | ||
|
Methylation in Case |
2.78E-01 (Median) | Methylation in Control | 6.32E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC22A17 in bladder cancer | [ 1 ] | |||
|
Location |
TSS1500 (cg09372750) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.63E+00 | Statistic Test | p-value:4.43E-07; Z-score:-6.54E+00 | ||
|
Methylation in Case |
2.58E-01 (Median) | Methylation in Control | 4.22E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC22A17 in colorectal cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg26922038) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:3.61E-06; Z-score:-2.05E+00 | ||
|
Methylation in Case |
7.65E-01 (Median) | Methylation in Control | 8.15E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC22A17 in hepatocellular carcinoma | [ 3 ] | |||
|
Location |
TSS1500 (cg09372750) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:1.64E-09; Z-score:-2.96E+00 | ||
|
Methylation in Case |
4.86E-01 (Median) | Methylation in Control | 6.56E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC22A17 in hepatocellular carcinoma | [ 3 ] | |||
|
Location |
Body (cg11817038) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.19E+00 | Statistic Test | p-value:9.13E-11; Z-score:-4.06E+00 | ||
|
Methylation in Case |
6.74E-01 (Median) | Methylation in Control | 8.04E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC22A17 in lung adenocarcinoma | [ 4 ] | |||
|
Location |
TSS1500 (cg09372750) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.18E+00 | Statistic Test | p-value:3.72E-02; Z-score:1.87E+00 | ||
|
Methylation in Case |
7.18E-01 (Median) | Methylation in Control | 6.06E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Cutaneous melanoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Complex methylation in Cutaneous melanoma | [ 5 ] | |||
|
Location |
Promoter (hypo) & Intragenic (hyper) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | TCGA/GEO bioinformatics & experimental validation | ||
|
Related Molecular Changes | Down regulation ofSLC22A17 | Experiment Method | Bisulfite sequencing, 5-Aza treatment, qRT-PCR | ||
|
Studied Phenotype |
Cutaneous melanoma[ ICD-11:2C30] | ||||
|
Experimental Material |
Melanoma cell lines, CM patient tissues vs nevi | ||||
|
Additional Notes |
SLC22A17 shows promoter hypomethylation (associated with activation) and intragenic hypermethylation (correlating with better prognosis) in CM. Two intragenic methylation hotspots are clinically significant. 5-Azacytidine treatment confirms epigenetic regulation. | ||||
|
Gastrointestinal cancers |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Promoter hypomethylation in GI cancers | [ 6 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | TCGA/GTEx bioinformatics | ||
|
Related Molecular Changes | Down regulation ofSLC22A17 | Experiment Method | Bisulfite sequencing, qRT-PCR | ||
|
Studied Phenotype |
Gastrointestinal cancers | ||||
|
Experimental Material |
Pan-cancer datasets (TCGA) | ||||
|
Additional Notes |
SLC22A17 shows consistent downregulation in GI cancers with promoter hypomethylation negatively correlating with expression. Part of LCN2-SLC22A17-MMP9 network regulating iron metabolism in TME. | ||||
|
Liver cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant/significant/moderate hypomethylation of SLC22A17 in liver cancer disease than that in healthy individual/adjacent tissue/other disease section | ||||
Studied Phenotype |
Liver cancer [ICD-11:2C12] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.43E-07; Fold-change:-0.355334115; Z-score:-1.840486939 | ||||
The Methylation Level of Disease Section Compare with the Adjacent Tissue |
p-value:4.89E-14; Fold-change:-0.360417357; Z-score:-26.14994106 | ||||
The Methylation Level of Disease Section Compare with the Other Disease Section |
p-value:0.038631317; Fold-change:-0.232239586; Z-score:-0.860611077 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue adjacent to the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
DT methylation level in tissue other than the diseased tissue of patients
|
|||||
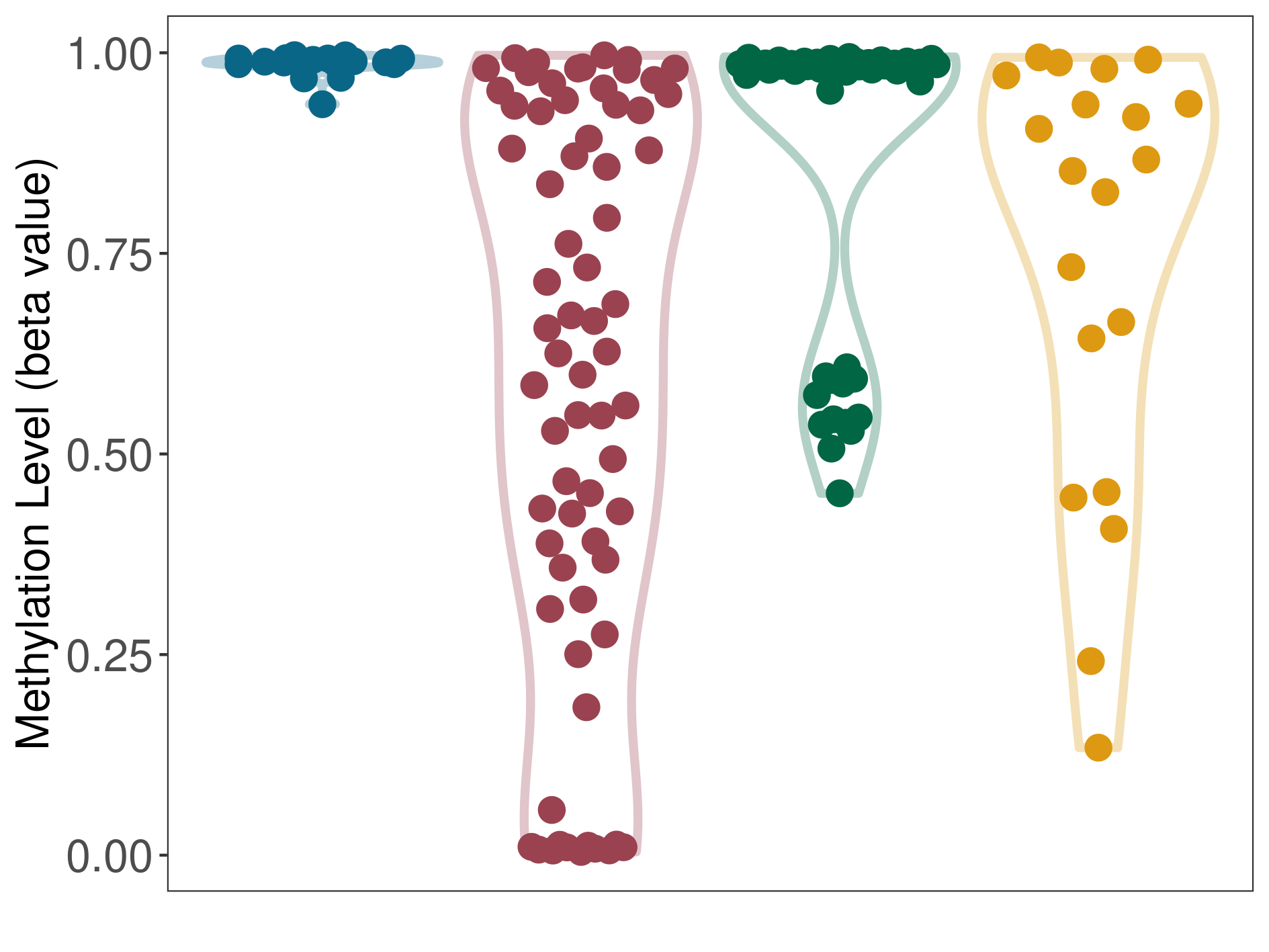
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Brain neuroblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC22A17 in brain neuroblastoma than that in healthy individual | ||||
Studied Phenotype |
Brain neuroblastoma [ICD-11:2A00.11] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:6.18E-21; Fold-change:-0.311157184; Z-score:-11.19061909 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
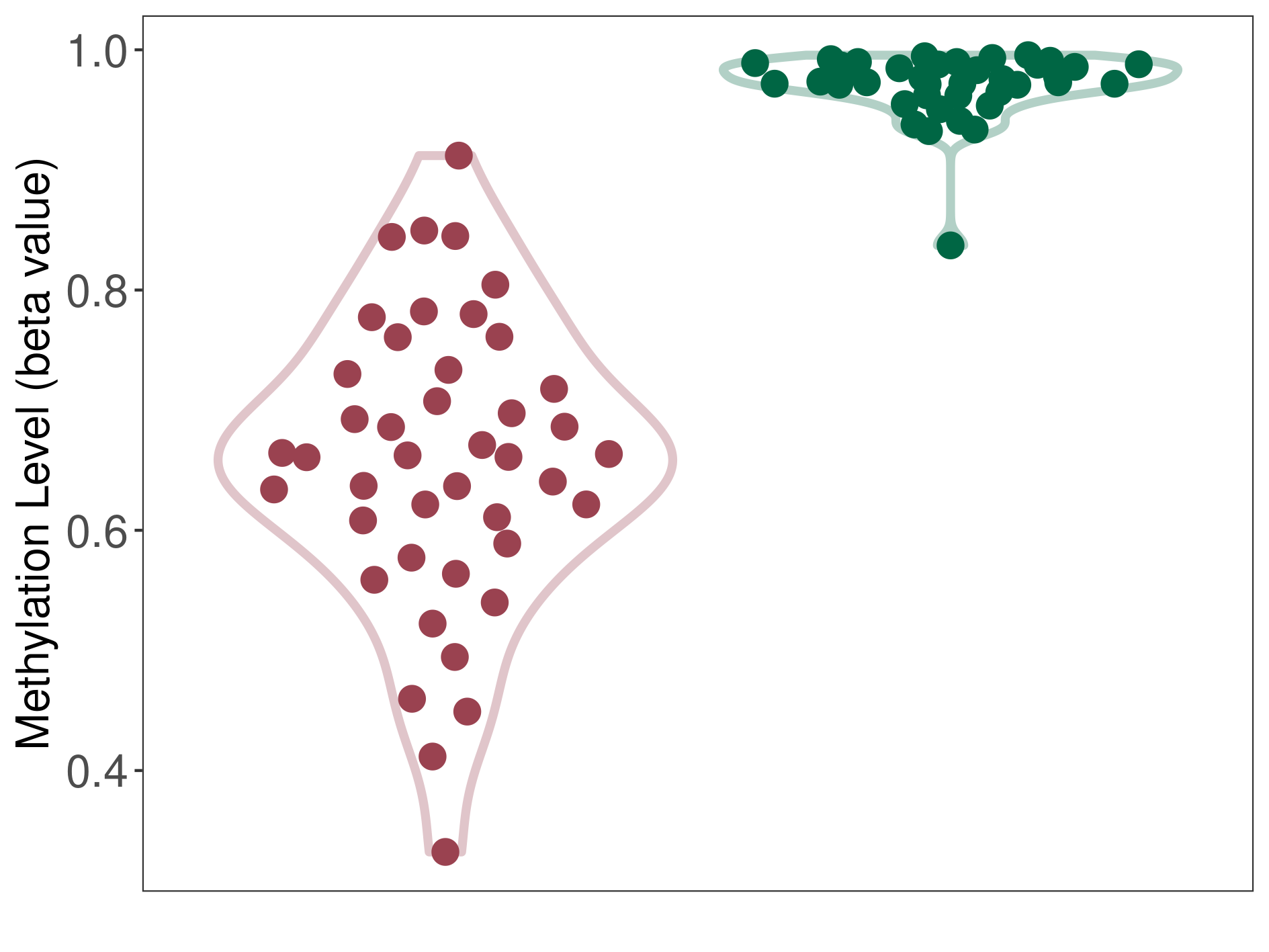
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
microRNA |
|||||
|
Unclear Phenotype |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
miR-3529 directly targets SLC22A17 | [ 7 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3529 | miRNA Mature ID | miR-3529-3p | ||
|
miRNA Sequence |
AACAACAAAAUCACUAGUCUUCCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon2 |
miR-5195 directly targets SLC22A17 | [ 7 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5195 | miRNA Mature ID | miR-5195-5p | ||
|
miRNA Sequence |
AACCCCUAAGGCAACUGGAUGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
