Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0128 Transporter Info | ||||
| Gene Name | SLC19A3 | ||||
| Transporter Name | Thiamine transporter 2 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Gastric cancer |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Hypermethylation of SLC19A3 in gastric cancer | [ 1 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Bisulfite sequencing | ||
|
Related Molecular Changes | Down regulation ofSLC19A3 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Gastric cancer[ ICD-11:2B72] | ||||
|
Frequency |
52 (51%) out of the 101 studied tumor samples | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
Hypermethylation within the SLC19A3 promoter is responsible for the down-regulation of SLC19A3 in gastric cancer. | ||||
|
Breast cancer |
13 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Hypermethylation of SLC19A3 in breast cancer | [ 2 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Methylation-specific PCR | ||
|
Related Molecular Changes | Down regulation ofSLC19A3 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Additional Notes |
SLC19A3 promoter Hypermethylation in plasma may be a novel biomarker for breast and gastric cancer diagnosis. | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC19A3 in breast cancer | [ 5 ] | |||
|
Location |
5'UTR (cg07417745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.83E+00 | Statistic Test | p-value:1.41E-17; Z-score:3.38E+00 | ||
|
Methylation in Case |
3.65E-01 (Median) | Methylation in Control | 1.99E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC19A3 in breast cancer | [ 5 ] | |||
|
Location |
5'UTR (cg25713185) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.31E+00 | Statistic Test | p-value:3.06E-13; Z-score:3.12E+00 | ||
|
Methylation in Case |
2.24E-01 (Median) | Methylation in Control | 9.70E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC19A3 in breast cancer | [ 5 ] | |||
|
Location |
TSS200 (cg19850080) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.52E+00 | Statistic Test | p-value:1.33E-07; Z-score:1.91E+00 | ||
|
Methylation in Case |
9.47E-02 (Median) | Methylation in Control | 6.21E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC19A3 in breast cancer | [ 5 ] | |||
|
Location |
TSS200 (cg01142776) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.45E+00 | Statistic Test | p-value:1.57E-07; Z-score:1.69E+00 | ||
|
Methylation in Case |
1.23E-01 (Median) | Methylation in Control | 8.46E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC19A3 in breast cancer | [ 5 ] | |||
|
Location |
TSS200 (cg02562740) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.51E+00 | Statistic Test | p-value:6.64E-04; Z-score:9.88E-01 | ||
|
Methylation in Case |
2.37E-02 (Median) | Methylation in Control | 1.57E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC19A3 in breast cancer | [ 5 ] | |||
|
Location |
TSS200 (cg20440901) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.86E+00 | Statistic Test | p-value:4.18E-03; Z-score:7.50E-01 | ||
|
Methylation in Case |
2.96E-02 (Median) | Methylation in Control | 1.59E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC19A3 in breast cancer | [ 5 ] | |||
|
Location |
TSS200 (cg05844583) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:2.89E-02; Z-score:1.21E-01 | ||
|
Methylation in Case |
6.61E-03 (Median) | Methylation in Control | 5.45E-03 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC19A3 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg11543686) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:8.55E-07; Z-score:-9.97E-01 | ||
|
Methylation in Case |
8.47E-01 (Median) | Methylation in Control | 8.77E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC19A3 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg19863633) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:7.72E-06; Z-score:-1.27E+00 | ||
|
Methylation in Case |
8.59E-01 (Median) | Methylation in Control | 8.91E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC19A3 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg04529897) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.95E-05; Z-score:-1.14E+00 | ||
|
Methylation in Case |
8.50E-01 (Median) | Methylation in Control | 8.90E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC19A3 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg21194765) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.09E-02; Z-score:-6.86E-01 | ||
|
Methylation in Case |
8.27E-01 (Median) | Methylation in Control | 8.60E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC19A3 in breast cancer | [ 5 ] | |||
|
Location |
3'UTR (cg04091914) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:3.81E-05; Z-score:-1.11E+00 | ||
|
Methylation in Case |
7.22E-01 (Median) | Methylation in Control | 8.02E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Atypical teratoid rhabdoid tumor |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC19A3 in atypical teratoid rhabdoid tumor | [ 3 ] | |||
|
Location |
5'UTR (cg03894284) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.44E+00 | Statistic Test | p-value:2.15E-08; Z-score:-1.18E+00 | ||
|
Methylation in Case |
3.75E-01 (Median) | Methylation in Control | 5.40E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC19A3 in atypical teratoid rhabdoid tumor | [ 3 ] | |||
|
Location |
5'UTR (cg07417745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:1.16E-07; Z-score:-1.32E+00 | ||
|
Methylation in Case |
8.28E-01 (Median) | Methylation in Control | 9.06E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC19A3 in atypical teratoid rhabdoid tumor | [ 3 ] | |||
|
Location |
5'UTR (cg13774987) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.16E+00 | Statistic Test | p-value:6.50E-07; Z-score:9.90E-01 | ||
|
Methylation in Case |
8.20E-01 (Median) | Methylation in Control | 7.10E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC19A3 in atypical teratoid rhabdoid tumor | [ 3 ] | |||
|
Location |
5'UTR (cg25713185) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.07E-05; Z-score:-1.22E+00 | ||
|
Methylation in Case |
8.59E-01 (Median) | Methylation in Control | 8.96E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC19A3 in atypical teratoid rhabdoid tumor | [ 3 ] | |||
|
Location |
Body (cg04529897) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.07E+00 | Statistic Test | p-value:8.56E-04; Z-score:-1.27E+00 | ||
|
Methylation in Case |
2.37E-01 (Median) | Methylation in Control | 4.90E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC19A3 in atypical teratoid rhabdoid tumor | [ 3 ] | |||
|
Location |
Body (cg11543686) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:2.29E-02; Z-score:4.44E-01 | ||
|
Methylation in Case |
9.08E-01 (Median) | Methylation in Control | 8.38E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC19A3 in atypical teratoid rhabdoid tumor | [ 3 ] | |||
|
Location |
3'UTR (cg04091914) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.06E+00 | Statistic Test | p-value:2.40E-14; Z-score:2.26E+00 | ||
|
Methylation in Case |
6.06E-01 (Median) | Methylation in Control | 2.94E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
15 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC19A3 in bladder cancer | [ 4 ] | |||
|
Location |
5'UTR (cg03894284) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:1.79E-04; Z-score:-2.74E+00 | ||
|
Methylation in Case |
5.99E-01 (Median) | Methylation in Control | 7.30E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC19A3 in bladder cancer | [ 4 ] | |||
|
Location |
5'UTR (cg25713185) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.94E+00 | Statistic Test | p-value:3.30E-04; Z-score:1.61E+01 | ||
|
Methylation in Case |
3.22E-01 (Median) | Methylation in Control | 1.10E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC19A3 in bladder cancer | [ 4 ] | |||
|
Location |
5'UTR (cg07417745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.10E+00 | Statistic Test | p-value:6.97E-04; Z-score:1.64E+01 | ||
|
Methylation in Case |
4.59E-01 (Median) | Methylation in Control | 2.19E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC19A3 in bladder cancer | [ 4 ] | |||
|
Location |
5'UTR (cg13774987) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:2.77E-02; Z-score:2.71E+00 | ||
|
Methylation in Case |
7.82E-01 (Median) | Methylation in Control | 6.82E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC19A3 in bladder cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg09726804) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:4.10E-02; Z-score:-1.25E+00 | ||
|
Methylation in Case |
7.69E-01 (Median) | Methylation in Control | 7.97E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC19A3 in bladder cancer | [ 4 ] | |||
|
Location |
TSS200 (cg19850080) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.76E+00 | Statistic Test | p-value:4.35E-03; Z-score:7.61E+00 | ||
|
Methylation in Case |
9.53E-02 (Median) | Methylation in Control | 5.41E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC19A3 in bladder cancer | [ 4 ] | |||
|
Location |
TSS200 (cg01142776) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.26E+00 | Statistic Test | p-value:1.03E-02; Z-score:3.00E+00 | ||
|
Methylation in Case |
1.03E-01 (Median) | Methylation in Control | 8.22E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC19A3 in bladder cancer | [ 4 ] | |||
|
Location |
TSS200 (cg20440901) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.85E+00 | Statistic Test | p-value:1.11E-02; Z-score:1.32E+00 | ||
|
Methylation in Case |
2.38E-02 (Median) | Methylation in Control | 1.29E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC19A3 in bladder cancer | [ 4 ] | |||
|
Location |
TSS200 (cg02562740) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.50E+00 | Statistic Test | p-value:2.10E-02; Z-score:1.90E+00 | ||
|
Methylation in Case |
2.35E-02 (Median) | Methylation in Control | 1.57E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC19A3 in bladder cancer | [ 4 ] | |||
|
Location |
TSS200 (cg05844583) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.59E+00 | Statistic Test | p-value:4.53E-02; Z-score:1.68E+00 | ||
|
Methylation in Case |
6.15E-03 (Median) | Methylation in Control | 2.37E-03 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC19A3 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg11543686) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:1.19E-04; Z-score:-9.66E+00 | ||
|
Methylation in Case |
8.08E-01 (Median) | Methylation in Control | 8.92E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC19A3 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg21194765) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.86E-03; Z-score:-4.95E+00 | ||
|
Methylation in Case |
8.65E-01 (Median) | Methylation in Control | 9.08E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC19A3 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg19863633) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:3.80E-03; Z-score:-2.47E+00 | ||
|
Methylation in Case |
8.80E-01 (Median) | Methylation in Control | 8.99E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC19A3 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg04529897) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.35E-03; Z-score:-1.19E+00 | ||
|
Methylation in Case |
8.88E-01 (Median) | Methylation in Control | 9.00E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC19A3 in bladder cancer | [ 4 ] | |||
|
Location |
3'UTR (cg04091914) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.20E+00 | Statistic Test | p-value:1.42E-04; Z-score:-6.85E+00 | ||
|
Methylation in Case |
6.79E-01 (Median) | Methylation in Control | 8.14E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC19A3 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
5'UTR (cg07417745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.65E+00 | Statistic Test | p-value:1.16E-04; Z-score:1.22E+00 | ||
|
Methylation in Case |
2.03E-01 (Median) | Methylation in Control | 1.23E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC19A3 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
5'UTR (cg25713185) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.08E+00 | Statistic Test | p-value:3.47E-04; Z-score:1.19E+00 | ||
|
Methylation in Case |
7.53E-02 (Median) | Methylation in Control | 3.62E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC19A3 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS200 (cg02562740) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:2.16E-07; Z-score:1.51E+00 | ||
|
Methylation in Case |
2.38E-02 (Median) | Methylation in Control | 2.02E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC19A3 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS200 (cg20440901) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:5.92E-06; Z-score:1.08E+00 | ||
|
Methylation in Case |
2.89E-02 (Median) | Methylation in Control | 2.57E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC19A3 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS200 (cg05844583) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:1.23E-05; Z-score:1.21E+00 | ||
|
Methylation in Case |
2.53E-02 (Median) | Methylation in Control | 2.23E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC19A3 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS200 (cg01142776) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.35E+00 | Statistic Test | p-value:1.15E-03; Z-score:8.58E-01 | ||
|
Methylation in Case |
4.99E-02 (Median) | Methylation in Control | 3.70E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC19A3 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS200 (cg19850080) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.18E+00 | Statistic Test | p-value:1.73E-03; Z-score:3.37E-01 | ||
|
Methylation in Case |
2.51E-02 (Median) | Methylation in Control | 2.14E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colon cancer |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC19A3 in colon adenocarcinoma | [ 7 ] | |||
|
Location |
5'UTR (cg01330016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.25E+00 | Statistic Test | p-value:8.38E-05; Z-score:-4.62E+00 | ||
|
Methylation in Case |
6.26E-01 (Median) | Methylation in Control | 7.85E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC19A3 in colon adenocarcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg01532168) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.32E+00 | Statistic Test | p-value:6.12E-07; Z-score:1.73E+00 | ||
|
Methylation in Case |
5.64E-01 (Median) | Methylation in Control | 4.26E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC19A3 in colon adenocarcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg10150176) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:2.15E-04; Z-score:-5.00E+00 | ||
|
Methylation in Case |
7.10E-01 (Median) | Methylation in Control | 8.25E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC19A3 in colon adenocarcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg04309194) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:3.07E-04; Z-score:-2.60E+00 | ||
|
Methylation in Case |
5.22E-01 (Median) | Methylation in Control | 6.08E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC19A3 in colon adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg16163535) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.21E+00 | Statistic Test | p-value:4.43E-06; Z-score:-3.68E+00 | ||
|
Methylation in Case |
6.96E-01 (Median) | Methylation in Control | 8.45E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC19A3 in colon adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg24642933) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.34E+00 | Statistic Test | p-value:8.55E-06; Z-score:-2.87E+00 | ||
|
Methylation in Case |
4.46E-01 (Median) | Methylation in Control | 5.98E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC19A3 in colon adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg08830588) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:3.47E-05; Z-score:-1.14E+00 | ||
|
Methylation in Case |
4.21E-01 (Median) | Methylation in Control | 4.87E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC19A3 in colorectal cancer | [ 8 ] | |||
|
Location |
5'UTR (cg13774987) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:3.42E-05; Z-score:1.19E+00 | ||
|
Methylation in Case |
9.13E-01 (Median) | Methylation in Control | 8.27E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC19A3 in colorectal cancer | [ 8 ] | |||
|
Location |
5'UTR (cg03894284) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.29E+00 | Statistic Test | p-value:4.47E-05; Z-score:1.38E+00 | ||
|
Methylation in Case |
8.19E-01 (Median) | Methylation in Control | 6.35E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC19A3 in colorectal cancer | [ 8 ] | |||
|
Location |
5'UTR (cg07417745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:1.70E-02; Z-score:3.85E-01 | ||
|
Methylation in Case |
3.42E-01 (Median) | Methylation in Control | 3.11E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC19A3 in colorectal cancer | [ 8 ] | |||
|
Location |
5'UTR (cg25713185) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:2.08E-02; Z-score:1.26E-01 | ||
|
Methylation in Case |
1.78E-01 (Median) | Methylation in Control | 1.70E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC19A3 in colorectal cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg09726804) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:1.34E-02; Z-score:7.24E-01 | ||
|
Methylation in Case |
8.98E-01 (Median) | Methylation in Control | 8.76E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC19A3 in colorectal cancer | [ 8 ] | |||
|
Location |
3'UTR (cg04091914) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:1.07E-02; Z-score:1.47E+00 | ||
|
Methylation in Case |
8.59E-01 (Median) | Methylation in Control | 8.20E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
11 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC19A3 in hepatocellular carcinoma | [ 9 ] | |||
|
Location |
5'UTR (cg03894284) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.38E+00 | Statistic Test | p-value:1.80E-06; Z-score:-1.20E+00 | ||
|
Methylation in Case |
3.59E-01 (Median) | Methylation in Control | 4.95E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC19A3 in hepatocellular carcinoma | [ 9 ] | |||
|
Location |
5'UTR (cg25713185) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.42E+00 | Statistic Test | p-value:1.94E-06; Z-score:-1.03E+00 | ||
|
Methylation in Case |
9.17E-02 (Median) | Methylation in Control | 1.31E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC19A3 in hepatocellular carcinoma | [ 9 ] | |||
|
Location |
5'UTR (cg07417745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.32E+00 | Statistic Test | p-value:2.25E-06; Z-score:-1.04E+00 | ||
|
Methylation in Case |
1.82E-01 (Median) | Methylation in Control | 2.41E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC19A3 in hepatocellular carcinoma | [ 9 ] | |||
|
Location |
TSS1500 (cg04981611) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:5.23E+00 | Statistic Test | p-value:3.54E-16; Z-score:1.50E+01 | ||
|
Methylation in Case |
3.46E-01 (Median) | Methylation in Control | 6.61E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC19A3 in hepatocellular carcinoma | [ 9 ] | |||
|
Location |
TSS200 (cg01142776) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:1.25E-06; Z-score:-1.16E+00 | ||
|
Methylation in Case |
7.82E-02 (Median) | Methylation in Control | 1.06E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC19A3 in hepatocellular carcinoma | [ 9 ] | |||
|
Location |
TSS200 (cg19850080) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.31E+00 | Statistic Test | p-value:3.41E-06; Z-score:-8.16E-01 | ||
|
Methylation in Case |
6.86E-02 (Median) | Methylation in Control | 8.95E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC19A3 in hepatocellular carcinoma | [ 9 ] | |||
|
Location |
Body (cg12303470) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.76E+00 | Statistic Test | p-value:2.68E-24; Z-score:-1.74E+01 | ||
|
Methylation in Case |
4.66E-01 (Median) | Methylation in Control | 8.20E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC19A3 in hepatocellular carcinoma | [ 9 ] | |||
|
Location |
Body (cg18869815) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.62E+00 | Statistic Test | p-value:1.58E-17; Z-score:-4.17E+00 | ||
|
Methylation in Case |
4.52E-01 (Median) | Methylation in Control | 7.33E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC19A3 in hepatocellular carcinoma | [ 9 ] | |||
|
Location |
Body (cg05118455) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.53E+00 | Statistic Test | p-value:5.12E-16; Z-score:-1.20E+01 | ||
|
Methylation in Case |
5.35E-01 (Median) | Methylation in Control | 8.17E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC19A3 in hepatocellular carcinoma | [ 9 ] | |||
|
Location |
Body (cg19863633) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.16E-02; Z-score:-5.83E-01 | ||
|
Methylation in Case |
8.78E-01 (Median) | Methylation in Control | 8.89E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC19A3 in hepatocellular carcinoma | [ 9 ] | |||
|
Location |
Body (cg21194765) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:3.97E-02; Z-score:-6.88E-02 | ||
|
Methylation in Case |
8.80E-01 (Median) | Methylation in Control | 8.81E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
HIV infection |
8 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC19A3 in HIV infection | [ 10 ] | |||
|
Location |
5'UTR (cg25713185) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.28E+00 | Statistic Test | p-value:3.04E-06; Z-score:1.56E+00 | ||
|
Methylation in Case |
2.63E-01 (Median) | Methylation in Control | 2.05E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC19A3 in HIV infection | [ 10 ] | |||
|
Location |
5'UTR (cg13774987) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:1.11E-03; Z-score:9.42E-01 | ||
|
Methylation in Case |
9.06E-01 (Median) | Methylation in Control | 8.87E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC19A3 in HIV infection | [ 10 ] | |||
|
Location |
5'UTR (cg07417745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:2.91E-02; Z-score:2.99E-01 | ||
|
Methylation in Case |
4.75E-01 (Median) | Methylation in Control | 4.60E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC19A3 in HIV infection | [ 10 ] | |||
|
Location |
TSS200 (cg01142776) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.37E+00 | Statistic Test | p-value:3.94E-05; Z-score:1.53E+00 | ||
|
Methylation in Case |
1.60E-01 (Median) | Methylation in Control | 1.17E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC19A3 in HIV infection | [ 10 ] | |||
|
Location |
TSS200 (cg19850080) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.40E+00 | Statistic Test | p-value:1.02E-04; Z-score:1.57E+00 | ||
|
Methylation in Case |
1.46E-01 (Median) | Methylation in Control | 1.04E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC19A3 in HIV infection | [ 10 ] | |||
|
Location |
TSS200 (cg02562740) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.49E+00 | Statistic Test | p-value:2.27E-03; Z-score:1.07E+00 | ||
|
Methylation in Case |
3.66E-02 (Median) | Methylation in Control | 2.45E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC19A3 in HIV infection | [ 10 ] | |||
|
Location |
TSS200 (cg20440901) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.31E+00 | Statistic Test | p-value:2.77E-02; Z-score:7.38E-01 | ||
|
Methylation in Case |
4.34E-02 (Median) | Methylation in Control | 3.32E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC19A3 in HIV infection | [ 10 ] | |||
|
Location |
3'UTR (cg04091914) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:5.76E-03; Z-score:-8.46E-01 | ||
|
Methylation in Case |
8.23E-01 (Median) | Methylation in Control | 8.62E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC19A3 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
5'UTR (cg03894284) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:1.40E-04; Z-score:2.36E+00 | ||
|
Methylation in Case |
8.39E-01 (Median) | Methylation in Control | 7.02E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC19A3 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
5'UTR (cg25713185) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.62E+00 | Statistic Test | p-value:6.22E-04; Z-score:3.35E+00 | ||
|
Methylation in Case |
2.88E-01 (Median) | Methylation in Control | 1.77E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC19A3 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
5'UTR (cg07417745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.50E+00 | Statistic Test | p-value:1.28E-03; Z-score:3.40E+00 | ||
|
Methylation in Case |
4.23E-01 (Median) | Methylation in Control | 2.81E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC19A3 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
5'UTR (cg13774987) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:7.73E-03; Z-score:1.83E+00 | ||
|
Methylation in Case |
8.41E-01 (Median) | Methylation in Control | 7.79E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC19A3 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
TSS200 (cg19850080) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.53E+00 | Statistic Test | p-value:5.21E-03; Z-score:3.09E+00 | ||
|
Methylation in Case |
1.59E-01 (Median) | Methylation in Control | 1.03E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC19A3 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
TSS200 (cg01142776) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.45E+00 | Statistic Test | p-value:7.40E-03; Z-score:4.20E+00 | ||
|
Methylation in Case |
1.71E-01 (Median) | Methylation in Control | 1.18E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Pancretic ductal adenocarcinoma |
12 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC19A3 in pancretic ductal adenocarcinoma | [ 12 ] | |||
|
Location |
5'UTR (cg20632143) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.18E+00 | Statistic Test | p-value:4.47E-08; Z-score:9.47E-01 | ||
|
Methylation in Case |
4.55E-01 (Median) | Methylation in Control | 3.85E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC19A3 in pancretic ductal adenocarcinoma | [ 12 ] | |||
|
Location |
TSS1500 (cg07845390) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.96E+00 | Statistic Test | p-value:1.76E-23; Z-score:4.95E+00 | ||
|
Methylation in Case |
2.43E-01 (Median) | Methylation in Control | 8.22E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC19A3 in pancretic ductal adenocarcinoma | [ 12 ] | |||
|
Location |
TSS1500 (cg22935149) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.31E+00 | Statistic Test | p-value:4.98E-14; Z-score:8.68E-01 | ||
|
Methylation in Case |
6.65E-02 (Median) | Methylation in Control | 5.09E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC19A3 in pancretic ductal adenocarcinoma | [ 12 ] | |||
|
Location |
1stExon (cg26923490) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.35E+00 | Statistic Test | p-value:8.88E-23; Z-score:3.94E+00 | ||
|
Methylation in Case |
4.72E-01 (Median) | Methylation in Control | 2.01E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC19A3 in pancretic ductal adenocarcinoma | [ 12 ] | |||
|
Location |
1stExon (cg01135800) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:4.05E-02; Z-score:-5.56E-01 | ||
|
Methylation in Case |
7.87E-01 (Median) | Methylation in Control | 8.32E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC19A3 in pancretic ductal adenocarcinoma | [ 12 ] | |||
|
Location |
Body (cg09565111) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:8.80E-10; Z-score:-1.44E+00 | ||
|
Methylation in Case |
6.73E-01 (Median) | Methylation in Control | 7.42E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC19A3 in pancretic ductal adenocarcinoma | [ 12 ] | |||
|
Location |
Body (cg26504422) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:9.10E-08; Z-score:1.92E+00 | ||
|
Methylation in Case |
8.24E-01 (Median) | Methylation in Control | 7.07E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC19A3 in pancretic ductal adenocarcinoma | [ 12 ] | |||
|
Location |
Body (cg09606015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.28E+00 | Statistic Test | p-value:3.00E-05; Z-score:-9.61E-01 | ||
|
Methylation in Case |
1.37E-01 (Median) | Methylation in Control | 1.75E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC19A3 in pancretic ductal adenocarcinoma | [ 12 ] | |||
|
Location |
Body (cg13971265) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:7.56E-04; Z-score:1.21E+00 | ||
|
Methylation in Case |
7.79E-01 (Median) | Methylation in Control | 7.07E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC19A3 in pancretic ductal adenocarcinoma | [ 12 ] | |||
|
Location |
Body (cg21627218) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:2.40E-03; Z-score:7.83E-01 | ||
|
Methylation in Case |
9.25E-01 (Median) | Methylation in Control | 9.17E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC19A3 in pancretic ductal adenocarcinoma | [ 12 ] | |||
|
Location |
Body (cg23761815) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.16E-02; Z-score:-3.74E-01 | ||
|
Methylation in Case |
8.81E-01 (Median) | Methylation in Control | 8.88E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC19A3 in pancretic ductal adenocarcinoma | [ 12 ] | |||
|
Location |
Body (cg18196463) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:3.09E-02; Z-score:-4.14E-01 | ||
|
Methylation in Case |
9.30E-01 (Median) | Methylation in Control | 9.35E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC19A3 in papillary thyroid cancer | [ 13 ] | |||
|
Location |
5'UTR (cg13774987) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:2.55E-03; Z-score:1.07E+00 | ||
|
Methylation in Case |
8.36E-01 (Median) | Methylation in Control | 7.58E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC19A3 in papillary thyroid cancer | [ 13 ] | |||
|
Location |
TSS200 (cg19850080) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:3.83E-03; Z-score:5.24E-01 | ||
|
Methylation in Case |
7.54E-02 (Median) | Methylation in Control | 6.80E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC19A3 in papillary thyroid cancer | [ 13 ] | |||
|
Location |
TSS200 (cg01142776) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:2.02E-02; Z-score:3.51E-01 | ||
|
Methylation in Case |
7.75E-02 (Median) | Methylation in Control | 7.15E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC19A3 in papillary thyroid cancer | [ 13 ] | |||
|
Location |
TSS200 (cg05844583) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:2.18E-02; Z-score:6.20E-01 | ||
|
Methylation in Case |
5.31E-02 (Median) | Methylation in Control | 4.97E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC19A3 in papillary thyroid cancer | [ 13 ] | |||
|
Location |
Body (cg11543686) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.20E-03; Z-score:-5.21E-01 | ||
|
Methylation in Case |
8.91E-01 (Median) | Methylation in Control | 9.02E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC19A3 in papillary thyroid cancer | [ 13 ] | |||
|
Location |
3'UTR (cg04091914) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.29E-03; Z-score:-1.84E-01 | ||
|
Methylation in Case |
8.59E-01 (Median) | Methylation in Control | 8.64E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC19A3 in prostate cancer | [ 14 ] | |||
|
Location |
TSS200 (cg02772823) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.97E+00 | Statistic Test | p-value:1.24E-02; Z-score:-2.78E+00 | ||
|
Methylation in Case |
2.86E-01 (Median) | Methylation in Control | 5.64E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC19A3 in prostate cancer | [ 14 ] | |||
|
Location |
Body (cg07986907) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:5.84E-03; Z-score:2.70E+00 | ||
|
Methylation in Case |
9.38E-01 (Median) | Methylation in Control | 8.16E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Moderate hypermethylation of SLC19A3 in prostate cancer than that in healthy individual | ||||
Studied Phenotype |
Prostate cancer [ICD-11:2C82] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.22E-34; Fold-change:0.233589324; Z-score:11.29860515 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
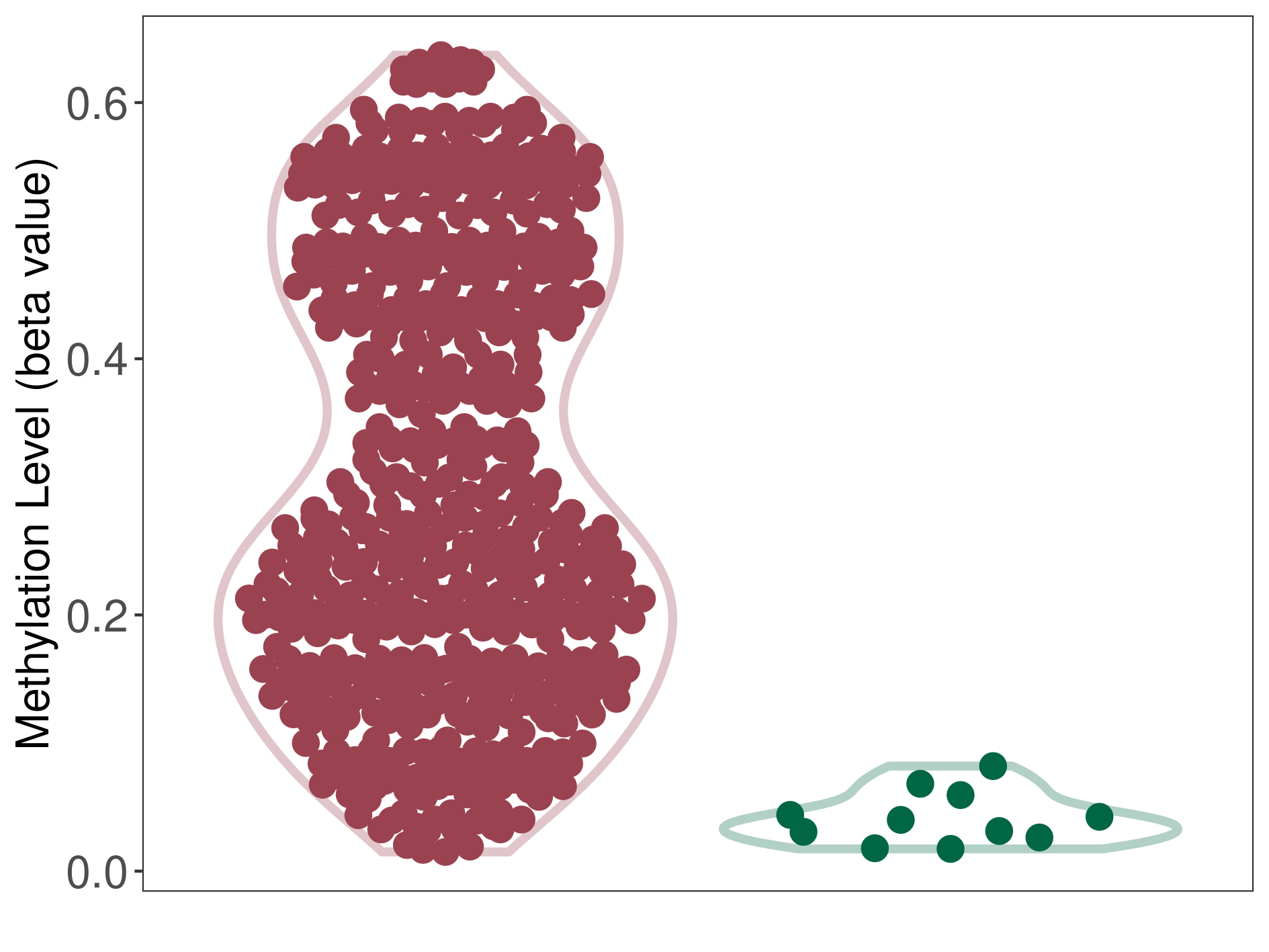
|
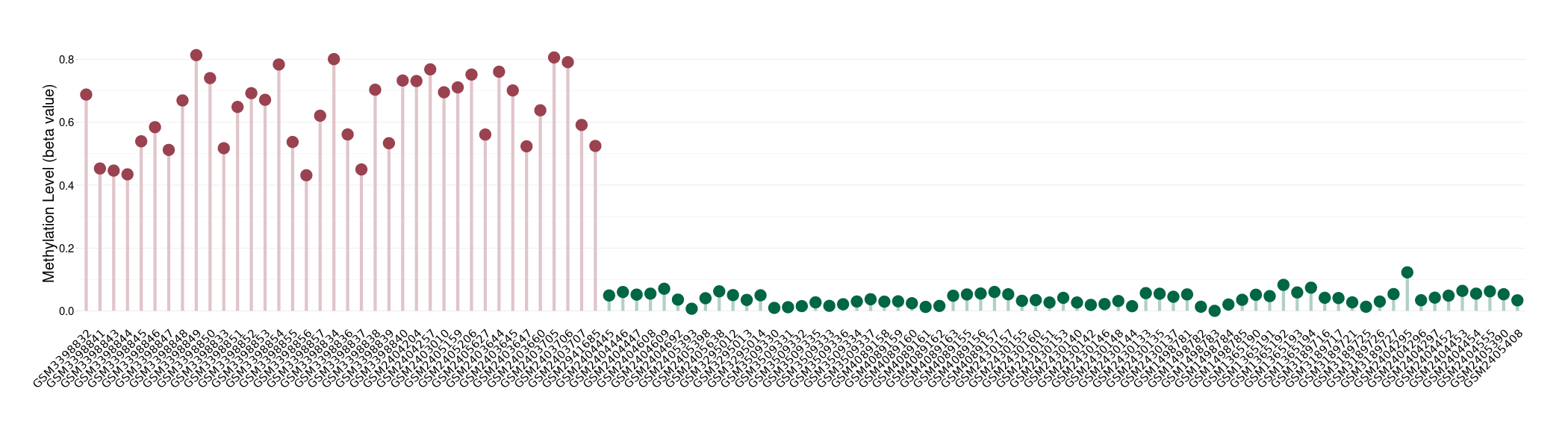
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Systemic lupus erythematosus |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC19A3 in systemic lupus erythematosus | [ 15 ] | |||
|
Location |
TSS200 (cg02562740) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:1.71E-02; Z-score:1.00E-01 | ||
|
Methylation in Case |
3.45E-02 (Median) | Methylation in Control | 3.35E-02 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC19A3 in systemic lupus erythematosus | [ 15 ] | |||
|
Location |
Body (cg21194765) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:6.12E-03; Z-score:-1.02E-01 | ||
|
Methylation in Case |
9.15E-01 (Median) | Methylation in Control | 9.17E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC19A3 in systemic lupus erythematosus | [ 15 ] | |||
|
Location |
Body (cg04529897) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:3.70E-02; Z-score:-9.31E-02 | ||
|
Methylation in Case |
9.20E-01 (Median) | Methylation in Control | 9.22E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Depression |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC19A3 in depression | [ 16 ] | |||
|
Location |
3'UTR (cg04091914) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:2.28E-02; Z-score:6.59E-01 | ||
|
Methylation in Case |
7.71E-01 (Median) | Methylation in Control | 7.53E-01 (Median) | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lymphoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
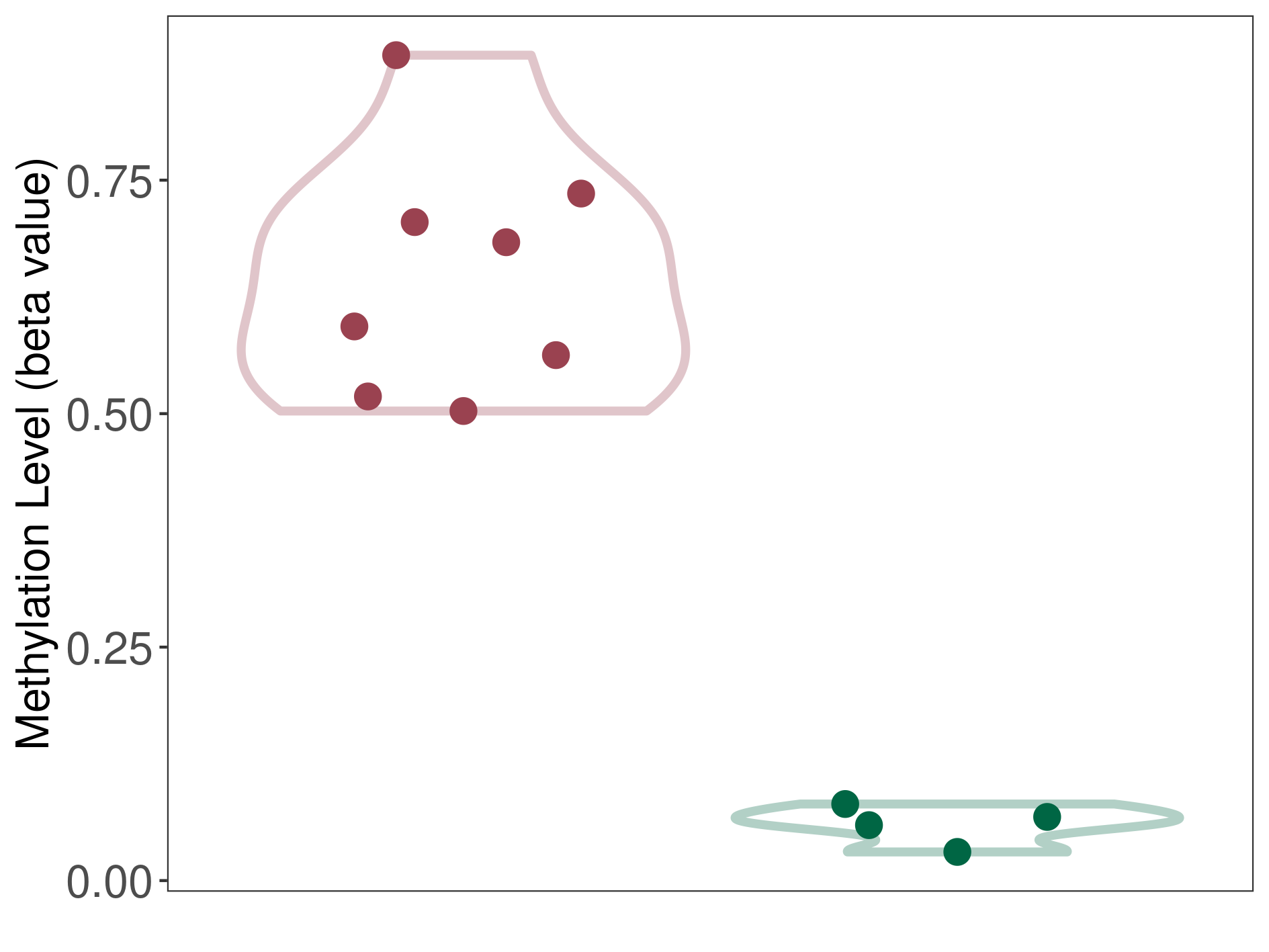
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC19A3 in lymphoma than that in healthy individual | ||||
Studied Phenotype |
Lymphoma [ICD-11:2B30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.63E-28; Fold-change:0.618985571; Z-score:29.85128227 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
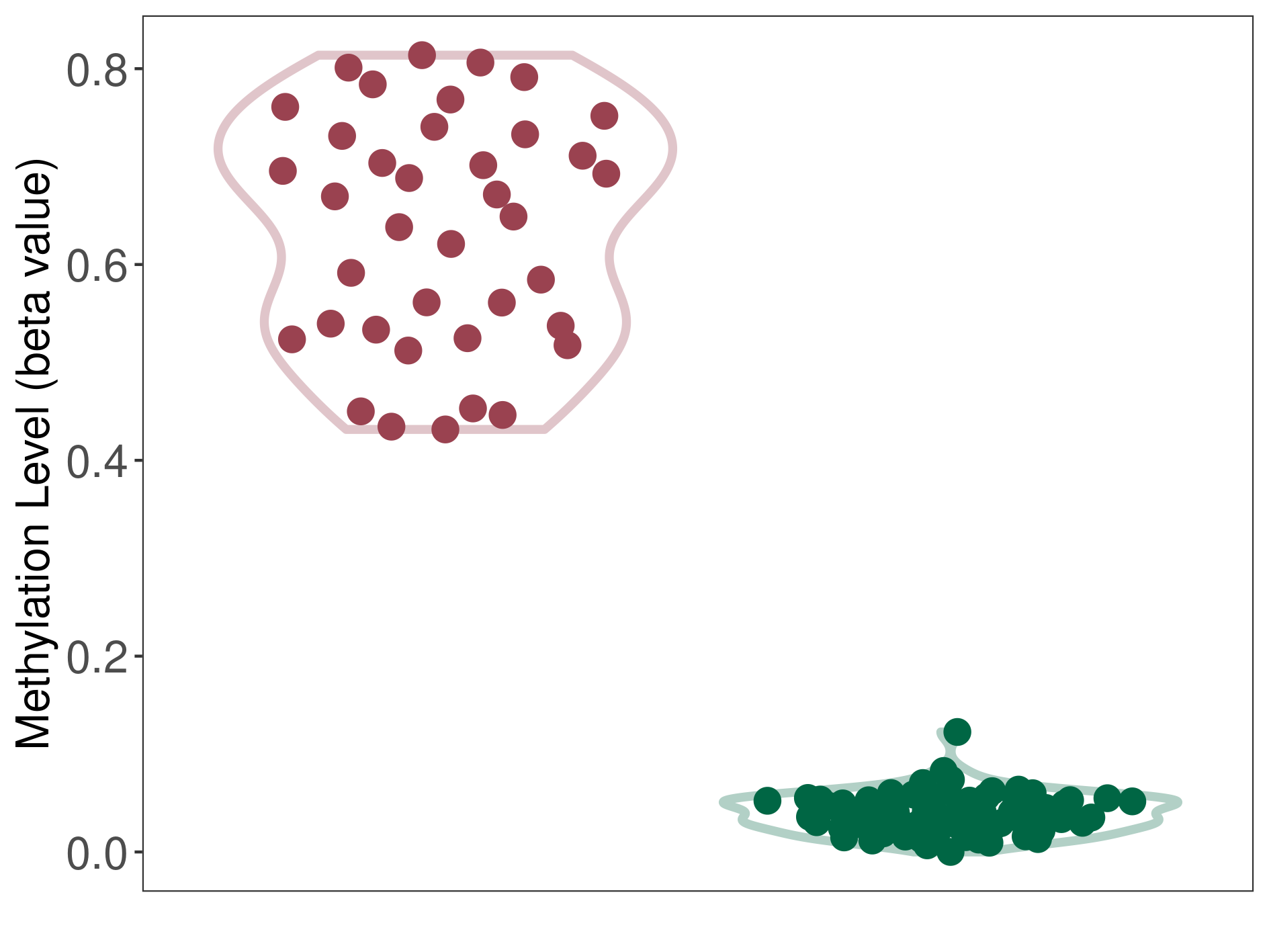
Prostate cancer metastasis |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC19A3 in prostate cancer metastasis than that in healthy individual | ||||
Studied Phenotype |
Prostate cancer metastasis [ICD-11:2.00E+06] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.96E-06; Fold-change:0.574690231; Z-score:26.50513269 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
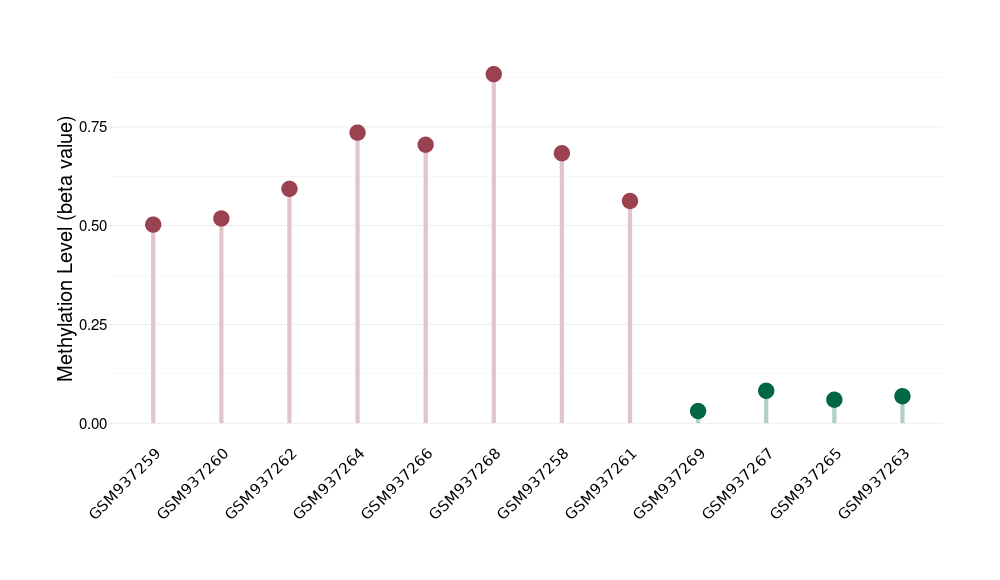 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Histone acetylation |
|||||
|
Colon cancer |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Hypoacetylation of SLC19A3 in colon cancer (compare with trichostatin A-treatment counterpart cells) | [ 17 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes | Down regulation ofSLC19A3 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
TSA treatment increased histone H3 acetylation of SLC19A3 promoter region in all 4 colon cancer cell lines examined. | ||||
|
Epigenetic Phenomenon2 |
Hypoacetylation of SLC19A3 in colon cancer (compare with TSA-treatment counterpart cells) | [ 17 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes | Down regulation ofSLC19A3 | Experiment Method | RT-qPCR | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Additional Notes |
In the 4 colon cancer cells examined, SLC19A3 mRNA expression is down-regulated by histone deacetylation. | ||||
|
microRNA |
|||||
|
Unclear Phenotype |
83 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
let-7a directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
let-7a | miRNA Mature ID | let-7a-5p | ||
|
miRNA Sequence |
UGAGGUAGUAGGUUGUAUAGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon2 |
let-7b directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
let-7b | miRNA Mature ID | let-7b-5p | ||
|
miRNA Sequence |
UGAGGUAGUAGGUUGUGUGGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon3 |
let-7c directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
let-7c | miRNA Mature ID | let-7c-5p | ||
|
miRNA Sequence |
UGAGGUAGUAGGUUGUAUGGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon4 |
let-7d directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
let-7d | miRNA Mature ID | let-7d-5p | ||
|
miRNA Sequence |
AGAGGUAGUAGGUUGCAUAGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon5 |
let-7e directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
let-7e | miRNA Mature ID | let-7e-5p | ||
|
miRNA Sequence |
UGAGGUAGGAGGUUGUAUAGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon6 |
let-7f directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
let-7f | miRNA Mature ID | let-7f-5p | ||
|
miRNA Sequence |
UGAGGUAGUAGAUUGUAUAGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon7 |
let-7g directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
let-7g | miRNA Mature ID | let-7g-5p | ||
|
miRNA Sequence |
UGAGGUAGUAGUUUGUACAGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon8 |
let-7i directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
let-7i | miRNA Mature ID | let-7i-5p | ||
|
miRNA Sequence |
UGAGGUAGUAGUUUGUGCUGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon9 |
miR-1273h directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-1273h | miRNA Mature ID | miR-1273h-5p | ||
|
miRNA Sequence |
CUGGGAGGUCAAGGCUGCAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon10 |
miR-1296 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1296 | miRNA Mature ID | miR-1296-3p | ||
|
miRNA Sequence |
GAGUGGGGCUUCGACCCUAACC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon11 |
miR-1343 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1343 | miRNA Mature ID | miR-1343-5p | ||
|
miRNA Sequence |
UGGGGAGCGGCCCCCGGGUGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon12 |
miR-135a directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-135a | miRNA Mature ID | miR-135a-5p | ||
|
miRNA Sequence |
UAUGGCUUUUUAUUCCUAUGUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon13 |
miR-135b directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-135b | miRNA Mature ID | miR-135b-5p | ||
|
miRNA Sequence |
UAUGGCUUUUCAUUCCUAUGUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon14 |
miR-149 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-149 | miRNA Mature ID | miR-149-3p | ||
|
miRNA Sequence |
AGGGAGGGACGGGGGCUGUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon15 |
miR-187 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-187 | miRNA Mature ID | miR-187-5p | ||
|
miRNA Sequence |
GGCUACAACACAGGACCCGGGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon16 |
miR-202 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-202 | miRNA Mature ID | miR-202-3p | ||
|
miRNA Sequence |
AGAGGUAUAGGGCAUGGGAA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon17 |
miR-203b directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-203b | miRNA Mature ID | miR-203b-3p | ||
|
miRNA Sequence |
UUGAACUGUUAAGAACCACUGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon18 |
miR-216a directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-216a | miRNA Mature ID | miR-216a-5p | ||
|
miRNA Sequence |
UAAUCUCAGCUGGCAACUGUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon19 |
miR-216b directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-216b | miRNA Mature ID | miR-216b-5p | ||
|
miRNA Sequence |
AAAUCUCUGCAGGCAAAUGUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon20 |
miR-221 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-221 | miRNA Mature ID | miR-221-3p | ||
|
miRNA Sequence |
AGCUACAUUGUCUGCUGGGUUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon21 |
miR-222 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-222 | miRNA Mature ID | miR-222-3p | ||
|
miRNA Sequence |
AGCUACAUCUGGCUACUGGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon22 |
miR-24 directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-24 | miRNA Mature ID | miR-24-3p | ||
|
miRNA Sequence |
UGGCUCAGUUCAGCAGGAACAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon23 |
miR-30b directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-30b | miRNA Mature ID | miR-30b-3p | ||
|
miRNA Sequence |
CUGGGAGGUGGAUGUUUACUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon24 |
miR-30c-1 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-30c-1 | miRNA Mature ID | miR-30c-1-3p | ||
|
miRNA Sequence |
CUGGGAGAGGGUUGUUUACUCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon25 |
miR-30c-2 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-30c-2 | miRNA Mature ID | miR-30c-2-3p | ||
|
miRNA Sequence |
CUGGGAGAAGGCUGUUUACUCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon26 |
miR-3122 directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3122 | miRNA Mature ID | miR-3122 | ||
|
miRNA Sequence |
GUUGGGACAAGAGGACGGUCUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon27 |
miR-3124 directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3124 | miRNA Mature ID | miR-3124-3p | ||
|
miRNA Sequence |
ACUUUCCUCACUCCCGUGAAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon28 |
miR-3150a directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3150a | miRNA Mature ID | miR-3150a-3p | ||
|
miRNA Sequence |
CUGGGGAGAUCCUCGAGGUUGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon29 |
miR-3175 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3175 | miRNA Mature ID | miR-3175 | ||
|
miRNA Sequence |
CGGGGAGAGAACGCAGUGACGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon30 |
miR-330 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-330 | miRNA Mature ID | miR-330-3p | ||
|
miRNA Sequence |
GCAAAGCACACGGCCUGCAGAGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon31 |
miR-3689a directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3689a | miRNA Mature ID | miR-3689a-3p | ||
|
miRNA Sequence |
CUGGGAGGUGUGAUAUCGUGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon32 |
miR-3689b directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3689b | miRNA Mature ID | miR-3689b-3p | ||
|
miRNA Sequence |
CUGGGAGGUGUGAUAUUGUGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon33 |
miR-3689c directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3689c | miRNA Mature ID | miR-3689c | ||
|
miRNA Sequence |
CUGGGAGGUGUGAUAUUGUGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon34 |
miR-3689d directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3689d | miRNA Mature ID | miR-3689d | ||
|
miRNA Sequence |
GGGAGGUGUGAUCUCACACUCG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon35 |
miR-371a directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-371a | miRNA Mature ID | miR-371a-5p | ||
|
miRNA Sequence |
ACUCAAACUGUGGGGGCACU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon36 |
miR-371b directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-371b | miRNA Mature ID | miR-371b-5p | ||
|
miRNA Sequence |
ACUCAAAAGAUGGCGGCACUUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon37 |
miR-372 directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-372 | miRNA Mature ID | miR-372-5p | ||
|
miRNA Sequence |
CCUCAAAUGUGGAGCACUAUUCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon38 |
miR-373 directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-373 | miRNA Mature ID | miR-373-5p | ||
|
miRNA Sequence |
ACUCAAAAUGGGGGCGCUUUCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon39 |
miR-3913 directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3913 | miRNA Mature ID | miR-3913-5p | ||
|
miRNA Sequence |
UUUGGGACUGAUCUUGAUGUCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon40 |
miR-3929 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3929 | miRNA Mature ID | miR-3929 | ||
|
miRNA Sequence |
GAGGCUGAUGUGAGUAGACCACU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon41 |
miR-4284 directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4284 | miRNA Mature ID | miR-4284 | ||
|
miRNA Sequence |
GGGCUCACAUCACCCCAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon42 |
miR-4311 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4311 | miRNA Mature ID | miR-4311 | ||
|
miRNA Sequence |
GAAAGAGAGCUGAGUGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon43 |
miR-4458 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4458 | miRNA Mature ID | miR-4458 | ||
|
miRNA Sequence |
AGAGGUAGGUGUGGAAGAA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon44 |
miR-4478 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4478 | miRNA Mature ID | miR-4478 | ||
|
miRNA Sequence |
GAGGCUGAGCUGAGGAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon45 |
miR-4481 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4481 | miRNA Mature ID | miR-4481 | ||
|
miRNA Sequence |
GGAGUGGGCUGGUGGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon46 |
miR-4500 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4500 | miRNA Mature ID | miR-4500 | ||
|
miRNA Sequence |
UGAGGUAGUAGUUUCUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon47 |
miR-4680 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4680 | miRNA Mature ID | miR-4680-5p | ||
|
miRNA Sequence |
AGAACUCUUGCAGUCUUAGAUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon48 |
miR-4693 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4693 | miRNA Mature ID | miR-4693-3p | ||
|
miRNA Sequence |
UGAGAGUGGAAUUCACAGUAUUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon49 |
miR-4698 directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4698 | miRNA Mature ID | miR-4698 | ||
|
miRNA Sequence |
UCAAAAUGUAGAGGAAGACCCCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon50 |
miR-4722 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4722 | miRNA Mature ID | miR-4722-5p | ||
|
miRNA Sequence |
GGCAGGAGGGCUGUGCCAGGUUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon51 |
miR-4728 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4728 | miRNA Mature ID | miR-4728-5p | ||
|
miRNA Sequence |
UGGGAGGGGAGAGGCAGCAAGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon52 |
miR-4745 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4745 | miRNA Mature ID | miR-4745-5p | ||
|
miRNA Sequence |
UGAGUGGGGCUCCCGGGACGGCG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon53 |
miR-4768 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4768 | miRNA Mature ID | miR-4768-3p | ||
|
miRNA Sequence |
CCAGGAGAUCCAGAGAGAAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon54 |
miR-485 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-485 | miRNA Mature ID | miR-485-5p | ||
|
miRNA Sequence |
AGAGGCUGGCCGUGAUGAAUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon55 |
miR-491 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-491 | miRNA Mature ID | miR-491-5p | ||
|
miRNA Sequence |
AGUGGGGAACCCUUCCAUGAGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon56 |
miR-548v directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-548v | miRNA Mature ID | miR-548v | ||
|
miRNA Sequence |
AGCUACAGUUACUUUUGCACCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon57 |
miR-616 directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-616 | miRNA Mature ID | miR-616-5p | ||
|
miRNA Sequence |
ACUCAAAACCCUUCAGUGACUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon58 |
miR-6165 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6165 | miRNA Mature ID | miR-6165 | ||
|
miRNA Sequence |
CAGCAGGAGGUGAGGGGAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon59 |
miR-6513 directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6513 | miRNA Mature ID | miR-6513-5p | ||
|
miRNA Sequence |
UUUGGGAUUGACGCCACAUGUCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon60 |
miR-665 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-665 | miRNA Mature ID | miR-665 | ||
|
miRNA Sequence |
ACCAGGAGGCUGAGGCCCCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon61 |
miR-6736 directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6736 | miRNA Mature ID | miR-6736-3p | ||
|
miRNA Sequence |
UCAGCUCCUCUCUACCCACAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon62 |
miR-6763 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6763 | miRNA Mature ID | miR-6763-5p | ||
|
miRNA Sequence |
CUGGGGAGUGGCUGGGGAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon63 |
miR-6779 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6779 | miRNA Mature ID | miR-6779-5p | ||
|
miRNA Sequence |
CUGGGAGGGGCUGGGUUUGGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon64 |
miR-6780a directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6780a | miRNA Mature ID | miR-6780a-5p | ||
|
miRNA Sequence |
UUGGGAGGGAAGACAGCUGGAGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon65 |
miR-6785 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6785 | miRNA Mature ID | miR-6785-5p | ||
|
miRNA Sequence |
UGGGAGGGCGUGGAUGAUGGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon66 |
miR-6787 directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6787 | miRNA Mature ID | miR-6787-3p | ||
|
miRNA Sequence |
UCUCAGCUGCUGCCCUCUCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon67 |
miR-6788 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6788 | miRNA Mature ID | miR-6788-5p | ||
|
miRNA Sequence |
CUGGGAGAAGAGUGGUGAAGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon68 |
miR-6799 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6799 | miRNA Mature ID | miR-6799-5p | ||
|
miRNA Sequence |
GGGGAGGUGUGCAGGGCUGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon69 |
miR-6825 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6825 | miRNA Mature ID | miR-6825-5p | ||
|
miRNA Sequence |
UGGGGAGGUGUGGAGUCAGCAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon70 |
miR-6851 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6851 | miRNA Mature ID | miR-6851-5p | ||
|
miRNA Sequence |
AGGAGGUGGUACUAGGGGCCAGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon71 |
miR-6883 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6883 | miRNA Mature ID | miR-6883-5p | ||
|
miRNA Sequence |
AGGGAGGGUGUGGUAUGGAUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon72 |
miR-6884 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6884 | miRNA Mature ID | miR-6884-5p | ||
|
miRNA Sequence |
AGAGGCUGAGAAGGUGAUGUUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon73 |
miR-6888 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6888 | miRNA Mature ID | miR-6888-5p | ||
|
miRNA Sequence |
AAGGAGAUGCUCAGGCAGAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon74 |
miR-7106 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-7106 | miRNA Mature ID | miR-7106-5p | ||
|
miRNA Sequence |
UGGGAGGAGGGGAUCUUGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon75 |
miR-7158 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-7158 | miRNA Mature ID | miR-7158-3p | ||
|
miRNA Sequence |
CUGAACUAGAGAUUGGGCCCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon76 |
miR-7160 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-7160 | miRNA Mature ID | miR-7160-5p | ||
|
miRNA Sequence |
UGCUGAGGUCCGGGCUGUGCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon77 |
miR-7162 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-7162 | miRNA Mature ID | miR-7162-3p | ||
|
miRNA Sequence |
UCUGAGGUGGAACAGCAGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon78 |
miR-8063 directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-8063 | miRNA Mature ID | miR-8063 | ||
|
miRNA Sequence |
UCAAAAUCAGGAGUCGGGGCUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon79 |
miR-887 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-887 | miRNA Mature ID | miR-887-5p | ||
|
miRNA Sequence |
CUUGGGAGCCCUGUUAGACUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon80 |
miR-890 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-890 | miRNA Mature ID | miR-890 | ||
|
miRNA Sequence |
UACUUGGAAAGGCAUCAGUUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon81 |
miR-9 directly targets SLC19A3 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-9 | miRNA Mature ID | miR-9-5p | ||
|
miRNA Sequence |
UCUUUGGUUAUCUAGCUGUAUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon82 |
miR-939 directly targets SLC19A3 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-939 | miRNA Mature ID | miR-939-5p | ||
|
miRNA Sequence |
UGGGGAGCUGAGGCUCUGGGGGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon83 |
miR-98 directly targets SLC19A3 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-98 | miRNA Mature ID | miR-98-5p | ||
|
miRNA Sequence |
UGAGGUAGUAAGUUGUAUUGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
