Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0122 Transporter Info | ||||
| Gene Name | SLC17A9 | ||||
| Transporter Name | Solute carrier family 17 member 9 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Pancretic ductal adenocarcinoma |
11 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC17A9 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg13766864) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:2.66E-05; Z-score:1.29E+00 | ||
|
Methylation in Case |
7.23E-01 (Median) | Methylation in Control | 6.19E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC17A9 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg20366258) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.32E+00 | Statistic Test | p-value:1.86E-04; Z-score:-9.13E-01 | ||
|
Methylation in Case |
1.47E-01 (Median) | Methylation in Control | 1.94E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC17A9 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg18219563) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:3.65E-09; Z-score:5.82E-01 | ||
|
Methylation in Case |
1.00E-01 (Median) | Methylation in Control | 8.83E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC17A9 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg21951425) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:4.26E+00 | Statistic Test | p-value:1.61E-06; Z-score:1.58E+00 | ||
|
Methylation in Case |
1.81E-01 (Median) | Methylation in Control | 4.24E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC17A9 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg25190638) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:4.36E-03; Z-score:6.14E-01 | ||
|
Methylation in Case |
8.49E-01 (Median) | Methylation in Control | 8.27E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC17A9 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
1stExon (cg20834681) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.34E+00 | Statistic Test | p-value:5.99E-11; Z-score:2.25E+00 | ||
|
Methylation in Case |
6.43E-01 (Median) | Methylation in Control | 4.78E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC17A9 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
1stExon (cg06723421) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:2.77E-04; Z-score:7.09E-01 | ||
|
Methylation in Case |
7.91E-01 (Median) | Methylation in Control | 7.75E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC17A9 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg16867817) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:6.88E-12; Z-score:-1.36E+00 | ||
|
Methylation in Case |
6.28E-01 (Median) | Methylation in Control | 6.87E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC17A9 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg23750338) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:3.95E-05; Z-score:-9.93E-01 | ||
|
Methylation in Case |
8.12E-01 (Median) | Methylation in Control | 8.34E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC17A9 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg14381448) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:1.44E-03; Z-score:-7.71E-01 | ||
|
Methylation in Case |
5.44E-01 (Median) | Methylation in Control | 5.91E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC17A9 in pancretic ductal adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg24585559) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.00E+00 | Statistic Test | p-value:8.86E-03; Z-score:4.30E-02 | ||
|
Methylation in Case |
8.46E-01 (Median) | Methylation in Control | 8.45E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
8 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC17A9 in prostate cancer | [ 2 ] | |||
|
Location |
5'UTR (cg00797102) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:3.97E-02; Z-score:1.17E+00 | ||
|
Methylation in Case |
8.33E-01 (Median) | Methylation in Control | 7.02E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC17A9 in prostate cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg25365006) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:3.35E-02; Z-score:1.69E+00 | ||
|
Methylation in Case |
9.13E-01 (Median) | Methylation in Control | 8.25E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC17A9 in prostate cancer | [ 2 ] | |||
|
Location |
TSS200 (cg07719512) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:4.93E-02; Z-score:2.76E+00 | ||
|
Methylation in Case |
7.75E-01 (Median) | Methylation in Control | 6.61E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC17A9 in prostate cancer | [ 2 ] | |||
|
Location |
Body (cg13681701) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.56E+00 | Statistic Test | p-value:2.39E-02; Z-score:-2.61E+00 | ||
|
Methylation in Case |
1.77E-01 (Median) | Methylation in Control | 2.75E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC17A9 in prostate cancer | [ 2 ] | |||
|
Location |
Body (cg08726801) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.51E+00 | Statistic Test | p-value:2.82E-02; Z-score:2.17E+00 | ||
|
Methylation in Case |
5.05E-01 (Median) | Methylation in Control | 3.35E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC17A9 in prostate cancer | [ 2 ] | |||
|
Location |
Body (cg10091752) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.54E+00 | Statistic Test | p-value:3.12E-02; Z-score:-7.24E+00 | ||
|
Methylation in Case |
2.77E-01 (Median) | Methylation in Control | 4.26E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC17A9 in prostate cancer | [ 2 ] | |||
|
Location |
Body (cg09958402) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:3.57E-02; Z-score:4.17E+00 | ||
|
Methylation in Case |
9.31E-01 (Median) | Methylation in Control | 8.72E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
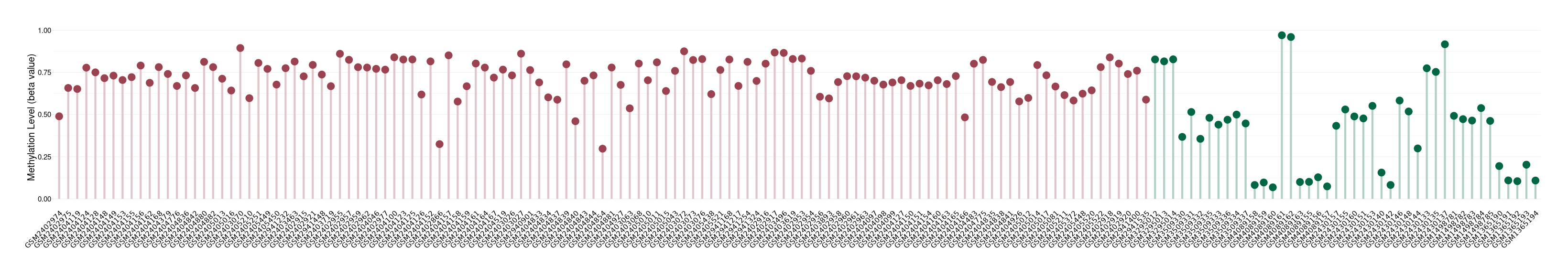
Epigenetic Phenomenon8 |
Significant hypermethylation of SLC17A9 in prostate cancer than that in healthy individual | ||||
Studied Phenotype |
Prostate cancer [ICD-11:2C82] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:2.47E-07; Fold-change:0.322979631; Z-score:4.100890028 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
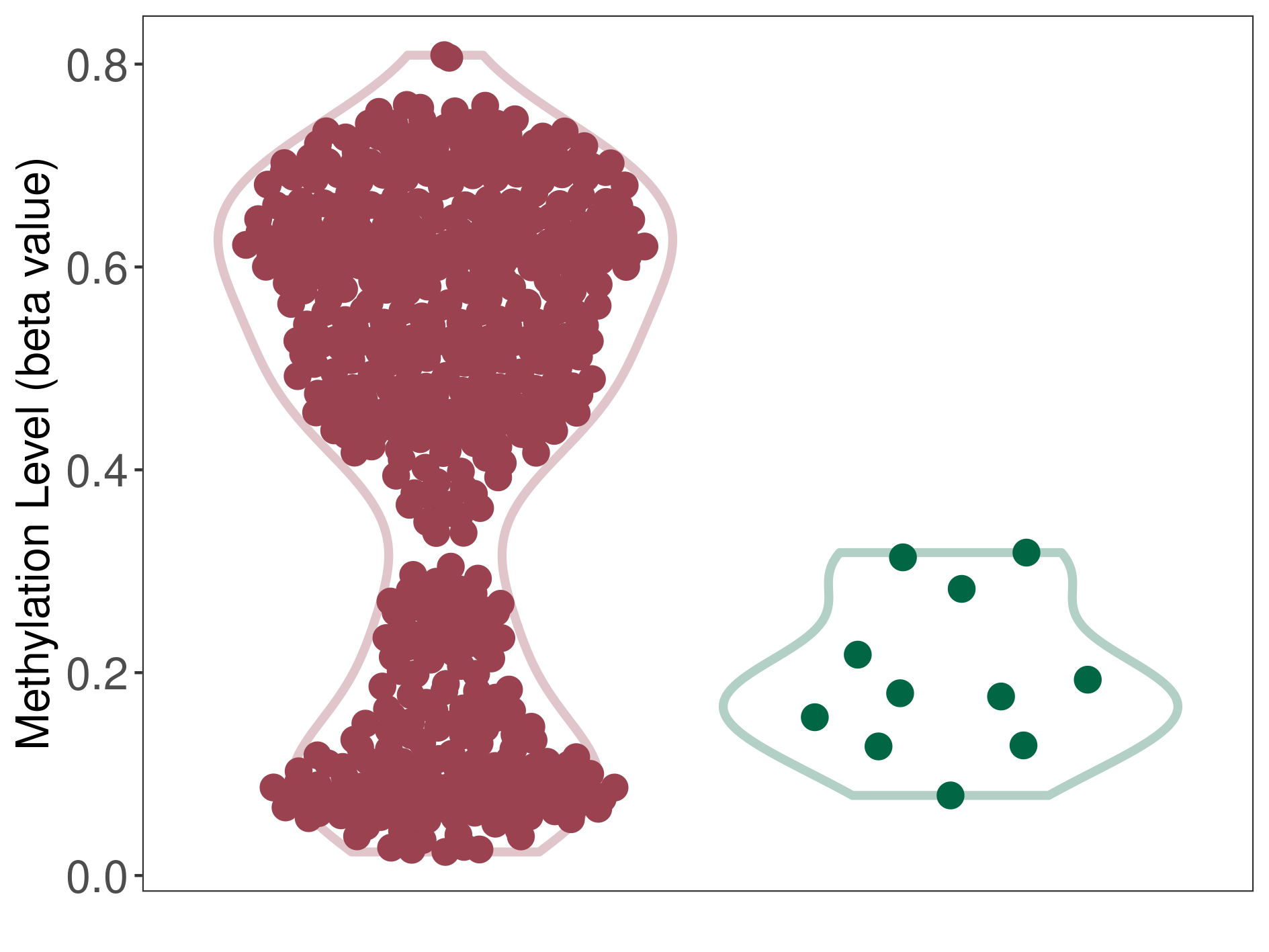
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Bladder cancer |
14 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC17A9 in bladder cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg22883889) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.53E+00 | Statistic Test | p-value:2.78E-08; Z-score:-6.28E+00 | ||
|
Methylation in Case |
2.66E-01 (Median) | Methylation in Control | 4.08E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC17A9 in bladder cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg21455983) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.27E+00 | Statistic Test | p-value:1.79E-07; Z-score:-1.57E+01 | ||
|
Methylation in Case |
6.50E-01 (Median) | Methylation in Control | 8.27E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC17A9 in bladder cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg00727912) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:8.04E-05; Z-score:-3.01E+00 | ||
|
Methylation in Case |
1.92E-01 (Median) | Methylation in Control | 2.62E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC17A9 in bladder cancer | [ 3 ] | |||
|
Location |
TSS200 (cg05534403) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.30E+00 | Statistic Test | p-value:1.16E-02; Z-score:-1.69E+00 | ||
|
Methylation in Case |
1.13E-01 (Median) | Methylation in Control | 1.46E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC17A9 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg07560932) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.61E+00 | Statistic Test | p-value:2.55E-11; Z-score:-1.69E+01 | ||
|
Methylation in Case |
5.41E-01 (Median) | Methylation in Control | 8.70E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC17A9 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg13044838) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.41E+00 | Statistic Test | p-value:1.99E-08; Z-score:-1.50E+01 | ||
|
Methylation in Case |
6.19E-01 (Median) | Methylation in Control | 8.74E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC17A9 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg12099423) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.88E+00 | Statistic Test | p-value:3.94E-07; Z-score:-9.33E+00 | ||
|
Methylation in Case |
3.49E-01 (Median) | Methylation in Control | 6.55E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC17A9 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg21627218) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:4.22E-07; Z-score:-4.31E+01 | ||
|
Methylation in Case |
6.80E-01 (Median) | Methylation in Control | 9.21E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC17A9 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg24866407) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.74E+00 | Statistic Test | p-value:1.11E-05; Z-score:-5.09E+00 | ||
|
Methylation in Case |
3.99E-01 (Median) | Methylation in Control | 6.95E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC17A9 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg19142181) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.25E+00 | Statistic Test | p-value:2.81E-04; Z-score:-3.84E+00 | ||
|
Methylation in Case |
5.69E-01 (Median) | Methylation in Control | 7.09E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC17A9 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg00688402) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:4.29E-04; Z-score:-3.04E+00 | ||
|
Methylation in Case |
5.41E-01 (Median) | Methylation in Control | 6.26E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC17A9 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg26733301) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.21E+00 | Statistic Test | p-value:6.57E-04; Z-score:1.19E+01 | ||
|
Methylation in Case |
2.29E-01 (Median) | Methylation in Control | 1.04E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC17A9 in bladder cancer | [ 3 ] | |||
|
Location |
Body (cg17221813) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:2.80E-02; Z-score:-6.30E-01 | ||
|
Methylation in Case |
7.33E-01 (Median) | Methylation in Control | 7.53E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC17A9 in bladder cancer | [ 3 ] | |||
|
Location |
3'UTR (cg13516768) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.54E+00 | Statistic Test | p-value:1.11E-07; Z-score:-8.39E+00 | ||
|
Methylation in Case |
5.68E-01 (Median) | Methylation in Control | 8.71E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
18 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg21455983) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:1.93E-03; Z-score:-8.63E-01 | ||
|
Methylation in Case |
8.01E-01 (Median) | Methylation in Control | 8.38E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg00727912) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:1.53E-02; Z-score:5.24E-01 | ||
|
Methylation in Case |
3.73E-01 (Median) | Methylation in Control | 3.46E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
TSS200 (cg07173972) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.43E+00 | Statistic Test | p-value:5.88E-10; Z-score:2.57E+00 | ||
|
Methylation in Case |
2.47E-01 (Median) | Methylation in Control | 1.02E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
TSS200 (cg26329715) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.08E+00 | Statistic Test | p-value:1.55E-09; Z-score:1.74E+00 | ||
|
Methylation in Case |
1.65E-01 (Median) | Methylation in Control | 7.91E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
TSS200 (cg00199007) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.02E+00 | Statistic Test | p-value:3.80E-09; Z-score:2.24E+00 | ||
|
Methylation in Case |
3.54E-01 (Median) | Methylation in Control | 1.76E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
TSS200 (cg16552454) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.96E+00 | Statistic Test | p-value:8.99E-09; Z-score:2.08E+00 | ||
|
Methylation in Case |
3.14E-01 (Median) | Methylation in Control | 1.60E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
TSS200 (cg05534403) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.27E+00 | Statistic Test | p-value:3.54E-06; Z-score:1.23E+00 | ||
|
Methylation in Case |
1.84E-01 (Median) | Methylation in Control | 1.45E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg26733301) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.88E+00 | Statistic Test | p-value:1.41E-10; Z-score:2.30E+00 | ||
|
Methylation in Case |
5.17E-01 (Median) | Methylation in Control | 2.75E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg13044838) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:2.42E-09; Z-score:-2.01E+00 | ||
|
Methylation in Case |
8.14E-01 (Median) | Methylation in Control | 8.76E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg24866407) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:4.87E-08; Z-score:1.75E+00 | ||
|
Methylation in Case |
7.42E-01 (Median) | Methylation in Control | 6.34E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg07560932) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:2.44E-06; Z-score:-1.54E+00 | ||
|
Methylation in Case |
7.83E-01 (Median) | Methylation in Control | 8.73E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg15677087) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:1.21E-05; Z-score:-1.16E+00 | ||
|
Methylation in Case |
7.09E-01 (Median) | Methylation in Control | 8.23E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg00688402) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:6.48E-04; Z-score:-1.20E+00 | ||
|
Methylation in Case |
6.03E-01 (Median) | Methylation in Control | 6.56E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg12099423) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:7.65E-04; Z-score:-1.41E+00 | ||
|
Methylation in Case |
5.63E-01 (Median) | Methylation in Control | 6.64E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg19142181) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:2.46E-03; Z-score:-9.02E-01 | ||
|
Methylation in Case |
6.87E-01 (Median) | Methylation in Control | 7.42E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg21627218) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:7.56E-03; Z-score:-5.75E-01 | ||
|
Methylation in Case |
9.03E-01 (Median) | Methylation in Control | 9.17E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
Body (cg17221813) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:2.49E-02; Z-score:-5.27E-01 | ||
|
Methylation in Case |
6.90E-01 (Median) | Methylation in Control | 7.60E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC17A9 in breast cancer | [ 4 ] | |||
|
Location |
3'UTR (cg13516768) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:1.55E-04; Z-score:-1.05E+00 | ||
|
Methylation in Case |
8.02E-01 (Median) | Methylation in Control | 8.64E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
8 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC17A9 in clear cell renal cell carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg22883889) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.21E+00 | Statistic Test | p-value:3.60E-06; Z-score:-2.75E+00 | ||
|
Methylation in Case |
5.09E-01 (Median) | Methylation in Control | 6.14E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC17A9 in clear cell renal cell carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg00727912) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:5.54E-03; Z-score:-1.48E+00 | ||
|
Methylation in Case |
3.43E-01 (Median) | Methylation in Control | 3.94E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC17A9 in clear cell renal cell carcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg16552454) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.57E+00 | Statistic Test | p-value:3.62E-06; Z-score:1.70E+00 | ||
|
Methylation in Case |
1.35E-01 (Median) | Methylation in Control | 5.28E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC17A9 in clear cell renal cell carcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg00199007) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.85E+00 | Statistic Test | p-value:2.06E-05; Z-score:1.23E+00 | ||
|
Methylation in Case |
1.31E-01 (Median) | Methylation in Control | 7.11E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC17A9 in clear cell renal cell carcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg26329715) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.53E+00 | Statistic Test | p-value:1.94E-04; Z-score:1.13E+00 | ||
|
Methylation in Case |
7.35E-02 (Median) | Methylation in Control | 4.82E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC17A9 in clear cell renal cell carcinoma | [ 5 ] | |||
|
Location |
Body (cg17221813) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:4.94E-06; Z-score:-1.47E+00 | ||
|
Methylation in Case |
8.20E-01 (Median) | Methylation in Control | 8.80E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC17A9 in clear cell renal cell carcinoma | [ 5 ] | |||
|
Location |
Body (cg12099423) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.55E-04; Z-score:-9.21E-01 | ||
|
Methylation in Case |
7.36E-01 (Median) | Methylation in Control | 7.79E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC17A9 in clear cell renal cell carcinoma | [ 5 ] | |||
|
Location |
Body (cg26733301) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.31E+00 | Statistic Test | p-value:1.60E-03; Z-score:1.07E+00 | ||
|
Methylation in Case |
2.20E-01 (Median) | Methylation in Control | 1.67E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colon cancer |
8 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC17A9 in colon adenocarcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg05581394) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.28E+00 | Statistic Test | p-value:3.37E-06; Z-score:-5.59E+00 | ||
|
Methylation in Case |
6.24E-01 (Median) | Methylation in Control | 7.97E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC17A9 in colon adenocarcinoma | [ 6 ] | |||
|
Location |
TSS200 (cg22113930) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.40E+00 | Statistic Test | p-value:4.06E-06; Z-score:1.27E+00 | ||
|
Methylation in Case |
6.48E-01 (Median) | Methylation in Control | 4.64E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC17A9 in colon adenocarcinoma | [ 6 ] | |||
|
Location |
1stExon (cg12052661) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.16E+00 | Statistic Test | p-value:1.21E-05; Z-score:1.95E+00 | ||
|
Methylation in Case |
6.26E-01 (Median) | Methylation in Control | 5.38E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC17A9 in colon adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg24712501) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.27E+00 | Statistic Test | p-value:3.38E-06; Z-score:-2.08E+00 | ||
|
Methylation in Case |
3.98E-01 (Median) | Methylation in Control | 5.05E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC17A9 in colon adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg15606745) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.17E+00 | Statistic Test | p-value:4.39E-05; Z-score:-2.18E+00 | ||
|
Methylation in Case |
5.28E-01 (Median) | Methylation in Control | 6.18E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC17A9 in colon adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg13603332) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:5.63E-05; Z-score:-2.60E+00 | ||
|
Methylation in Case |
4.97E-01 (Median) | Methylation in Control | 5.87E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC17A9 in colon adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg11034122) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:2.88E-04; Z-score:6.59E-01 | ||
|
Methylation in Case |
7.63E-01 (Median) | Methylation in Control | 7.13E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC17A9 in colon adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg20475114) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:8.78E-04; Z-score:-1.99E+00 | ||
|
Methylation in Case |
6.13E-01 (Median) | Methylation in Control | 6.79E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC17A9 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg21455983) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:2.53E-06; Z-score:-1.65E+00 | ||
|
Methylation in Case |
8.85E-01 (Median) | Methylation in Control | 9.12E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC17A9 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg22883889) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:9.51E-03; Z-score:-6.18E-01 | ||
|
Methylation in Case |
3.88E-01 (Median) | Methylation in Control | 4.48E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC17A9 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg15677087) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.47E+00 | Statistic Test | p-value:5.41E-08; Z-score:-1.98E+00 | ||
|
Methylation in Case |
5.57E-01 (Median) | Methylation in Control | 8.18E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC17A9 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg13044838) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:7.74E-04; Z-score:-9.12E-01 | ||
|
Methylation in Case |
9.06E-01 (Median) | Methylation in Control | 9.24E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC17A9 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg21627218) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:1.94E-03; Z-score:5.82E-01 | ||
|
Methylation in Case |
9.59E-01 (Median) | Methylation in Control | 9.53E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC17A9 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg26733301) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.45E+00 | Statistic Test | p-value:3.31E-02; Z-score:6.92E-01 | ||
|
Methylation in Case |
4.19E-01 (Median) | Methylation in Control | 2.89E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC17A9 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg07560932) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.00E+00 | Statistic Test | p-value:3.70E-02; Z-score:1.61E-01 | ||
|
Methylation in Case |
9.32E-01 (Median) | Methylation in Control | 9.29E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC17A9 in colorectal cancer | [ 7 ] | |||
|
Location |
3'UTR (cg13516768) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:1.25E-02; Z-score:1.03E+00 | ||
|
Methylation in Case |
7.64E-01 (Median) | Methylation in Control | 7.10E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
17 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC17A9 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS1500 (cg23986335) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.49E+00 | Statistic Test | p-value:3.04E-12; Z-score:5.46E+00 | ||
|
Methylation in Case |
2.48E-01 (Median) | Methylation in Control | 9.96E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC17A9 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS1500 (cg00727912) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:1.21E-05; Z-score:-1.15E+00 | ||
|
Methylation in Case |
1.92E-01 (Median) | Methylation in Control | 2.61E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC17A9 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS1500 (cg21455983) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.28E-03; Z-score:-1.54E-01 | ||
|
Methylation in Case |
7.94E-01 (Median) | Methylation in Control | 8.00E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC17A9 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg16552454) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.54E+00 | Statistic Test | p-value:1.09E-04; Z-score:-9.34E-01 | ||
|
Methylation in Case |
1.30E-01 (Median) | Methylation in Control | 2.01E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC17A9 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg00199007) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.48E+00 | Statistic Test | p-value:3.12E-04; Z-score:-8.44E-01 | ||
|
Methylation in Case |
1.39E-01 (Median) | Methylation in Control | 2.07E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC17A9 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg07173972) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.50E+00 | Statistic Test | p-value:8.08E-04; Z-score:-7.44E-01 | ||
|
Methylation in Case |
8.01E-02 (Median) | Methylation in Control | 1.20E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC17A9 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg05534403) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:9.00E-04; Z-score:-5.10E-01 | ||
|
Methylation in Case |
1.08E-01 (Median) | Methylation in Control | 1.23E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC17A9 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg26329715) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.37E+00 | Statistic Test | p-value:2.17E-03; Z-score:-6.32E-01 | ||
|
Methylation in Case |
7.15E-02 (Median) | Methylation in Control | 9.80E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC17A9 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg19969104) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.43E+00 | Statistic Test | p-value:1.03E-12; Z-score:-3.38E+00 | ||
|
Methylation in Case |
4.73E-01 (Median) | Methylation in Control | 6.75E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC17A9 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg15677087) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.23E+00 | Statistic Test | p-value:4.23E-08; Z-score:-1.18E+00 | ||
|
Methylation in Case |
4.40E-01 (Median) | Methylation in Control | 5.40E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC17A9 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg17221813) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:5.85E-06; Z-score:8.34E-01 | ||
|
Methylation in Case |
7.87E-01 (Median) | Methylation in Control | 7.27E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC17A9 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg12099423) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:2.50E-05; Z-score:8.99E-01 | ||
|
Methylation in Case |
7.14E-01 (Median) | Methylation in Control | 6.40E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC17A9 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg19142181) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:5.53E-04; Z-score:8.17E-01 | ||
|
Methylation in Case |
8.02E-01 (Median) | Methylation in Control | 7.55E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC17A9 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg07560932) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:8.21E-04; Z-score:-2.66E-01 | ||
|
Methylation in Case |
8.70E-01 (Median) | Methylation in Control | 8.75E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC17A9 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg24866407) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.19E-02; Z-score:-1.00E-01 | ||
|
Methylation in Case |
7.99E-01 (Median) | Methylation in Control | 8.05E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC17A9 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
3'UTR (cg12562245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.27E+00 | Statistic Test | p-value:4.43E-10; Z-score:-3.52E+00 | ||
|
Methylation in Case |
6.10E-01 (Median) | Methylation in Control | 7.76E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC17A9 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
3'UTR (cg13516768) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:2.17E-02; Z-score:-3.11E-01 | ||
|
Methylation in Case |
8.18E-01 (Median) | Methylation in Control | 8.32E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC17A9 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
TSS1500 (cg00727912) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:8.26E-05; Z-score:-1.15E+00 | ||
|
Methylation in Case |
3.69E-01 (Median) | Methylation in Control | 4.29E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC17A9 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
TSS200 (cg00199007) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.24E+00 | Statistic Test | p-value:2.20E-02; Z-score:-9.27E-01 | ||
|
Methylation in Case |
1.53E-01 (Median) | Methylation in Control | 1.89E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC17A9 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
TSS200 (cg16552454) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.19E+00 | Statistic Test | p-value:3.39E-02; Z-score:-5.18E-01 | ||
|
Methylation in Case |
1.34E-01 (Median) | Methylation in Control | 1.59E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC17A9 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
Body (cg15677087) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:6.21E-06; Z-score:-1.04E+00 | ||
|
Methylation in Case |
8.34E-01 (Median) | Methylation in Control | 8.82E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC17A9 in papillary thyroid cancer | [ 9 ] | |||
|
Location |
Body (cg26733301) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:1.09E-03; Z-score:-7.21E-01 | ||
|
Methylation in Case |
9.65E-02 (Median) | Methylation in Control | 1.14E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC17A9 in panic disorder | [ 10 ] | |||
|
Location |
TSS200 (cg00199007) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-8.56E-01 | Statistic Test | p-value:7.75E-03; Z-score:-4.23E-01 | ||
|
Methylation in Case |
-2.42E+00 (Median) | Methylation in Control | -2.07E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC17A9 in panic disorder | [ 10 ] | |||
|
Location |
TSS200 (cg16552454) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-9.22E-01 | Statistic Test | p-value:1.76E-02; Z-score:-2.60E-01 | ||
|
Methylation in Case |
-2.55E+00 (Median) | Methylation in Control | -2.35E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC17A9 in panic disorder | [ 10 ] | |||
|
Location |
Body (cg12099423) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:7.92E-01 | Statistic Test | p-value:3.26E-02; Z-score:2.23E-01 | ||
|
Methylation in Case |
-1.00E+00 (Median) | Methylation in Control | -1.26E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC17A9 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
Body (cg15677087) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.24E+00 | Statistic Test | p-value:2.13E-02; Z-score:-5.21E+00 | ||
|
Methylation in Case |
6.90E-01 (Median) | Methylation in Control | 8.53E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC17A9 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
Body (cg26733301) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.62E+00 | Statistic Test | p-value:2.50E-02; Z-score:8.76E+00 | ||
|
Methylation in Case |
2.64E-01 (Median) | Methylation in Control | 1.01E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC17A9 in lung adenocarcinoma | [ 11 ] | |||
|
Location |
3'UTR (cg13516768) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:4.34E-02; Z-score:-1.34E+00 | ||
|
Methylation in Case |
7.79E-01 (Median) | Methylation in Control | 8.23E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Atypical teratoid rhabdoid tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypermethylation of SLC17A9 in atypical teratoid rhabdoid tumour than that in healthy individual | ||||
Studied Phenotype |
Atypical teratoid rhabdoid tumour [ICD-11:2A00.1Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.03E-08; Fold-change:0.263880986; Z-score:0.982221418 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
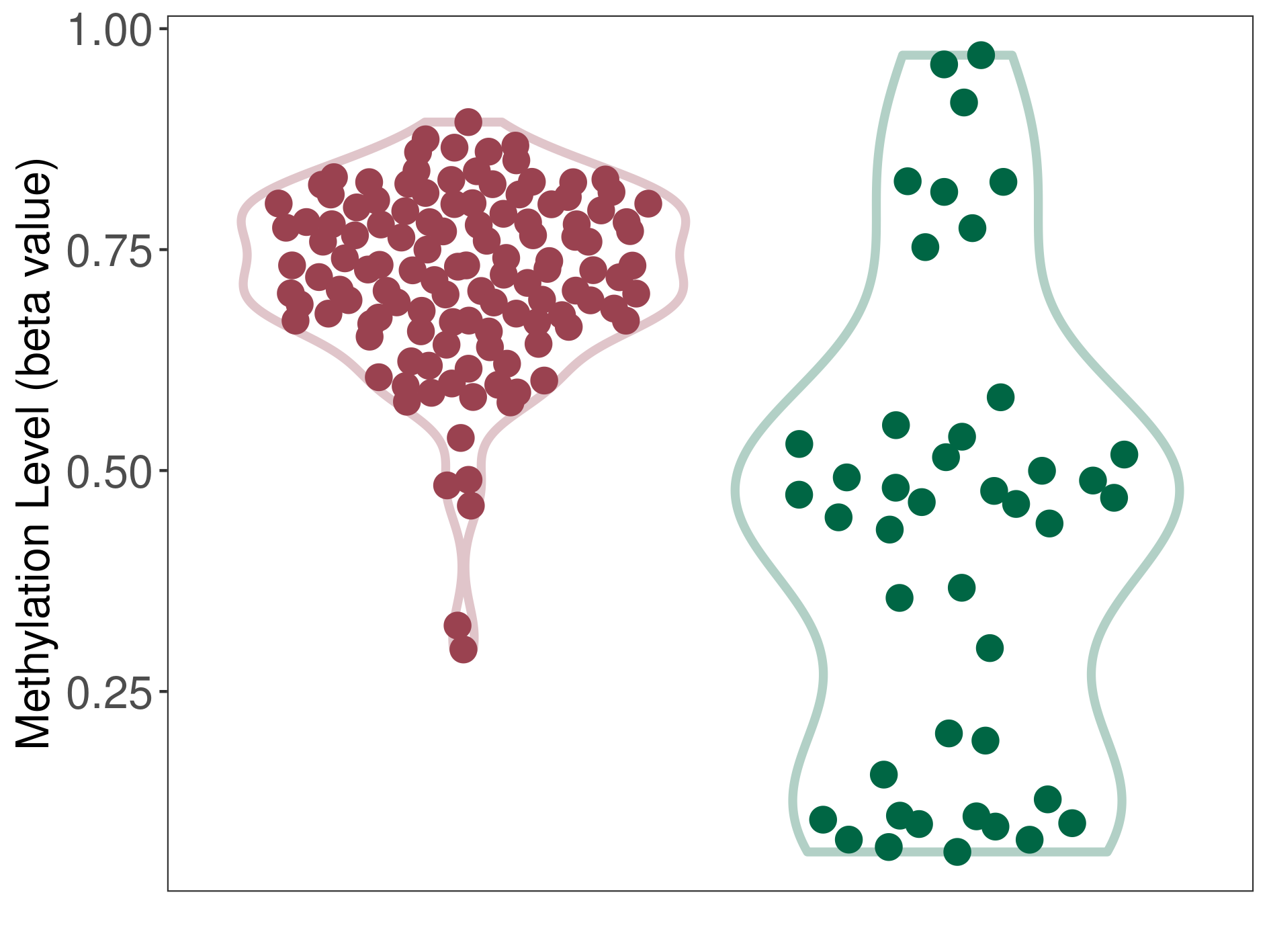
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Oligodendroglial tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypermethylation of SLC17A9 in oligodendroglial tumour than that in healthy individual | ||||
Studied Phenotype |
Oligodendroglial tumour [ICD-11:2A00.Y1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:6.39E-07; Fold-change:0.26472195; Z-score:8.285122349 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
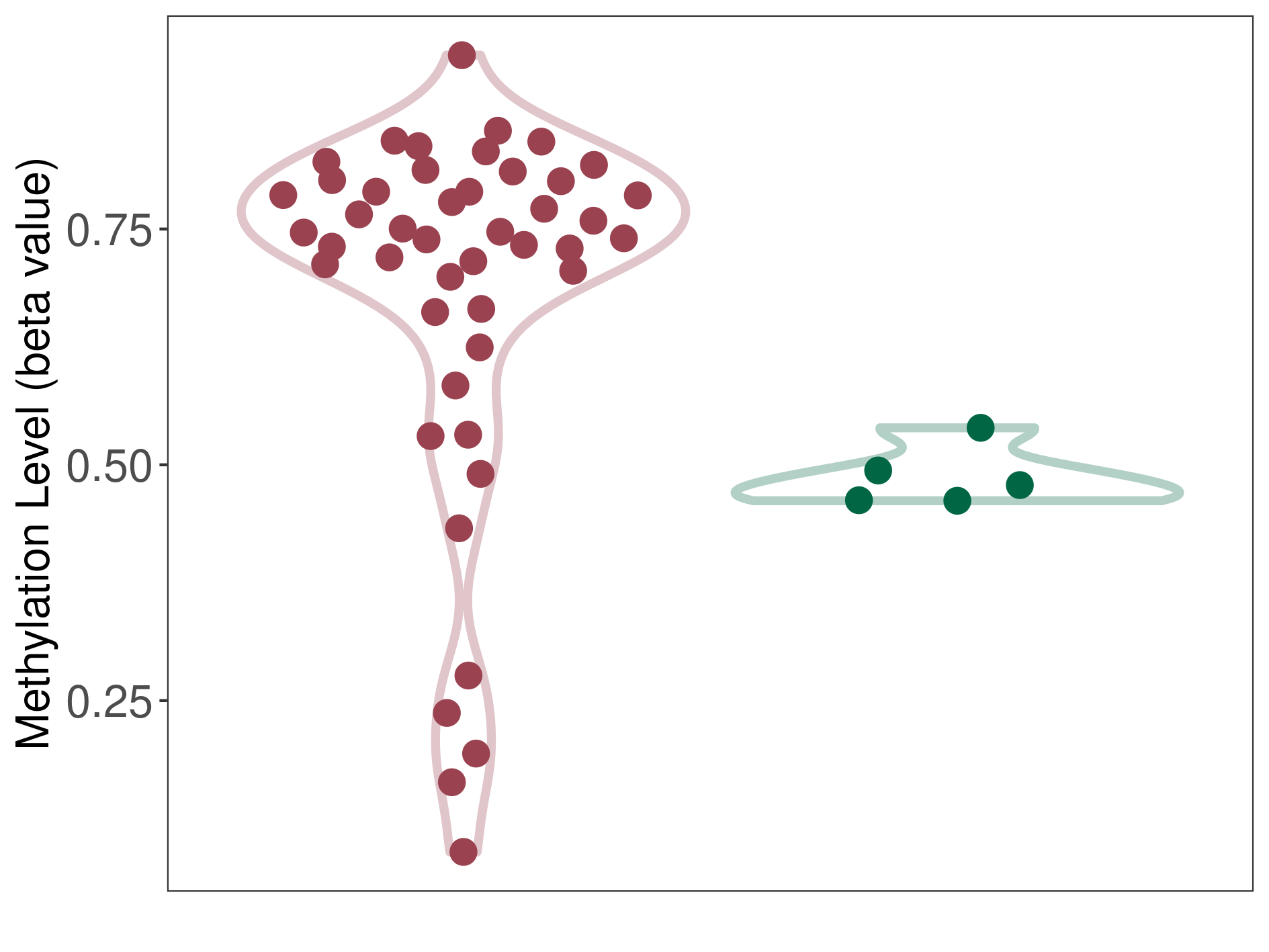
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
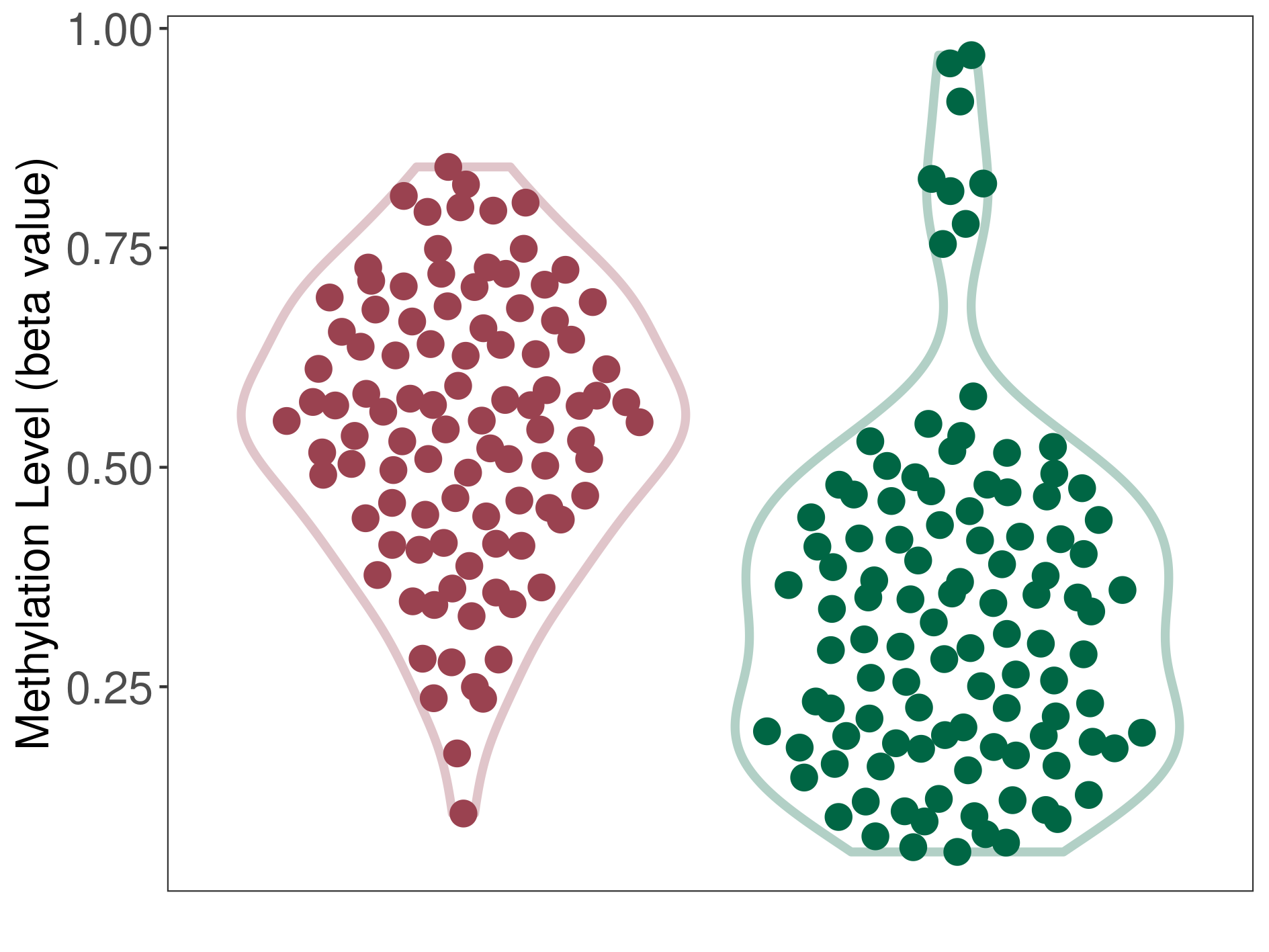
RELA YAP fusion ependymoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypermethylation of SLC17A9 in rela yap fusion ependymoma than that in healthy individual | ||||
Studied Phenotype |
RELA YAP fusion ependymoma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:4.65E-14; Fold-change:0.234980529; Z-score:1.173571586 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
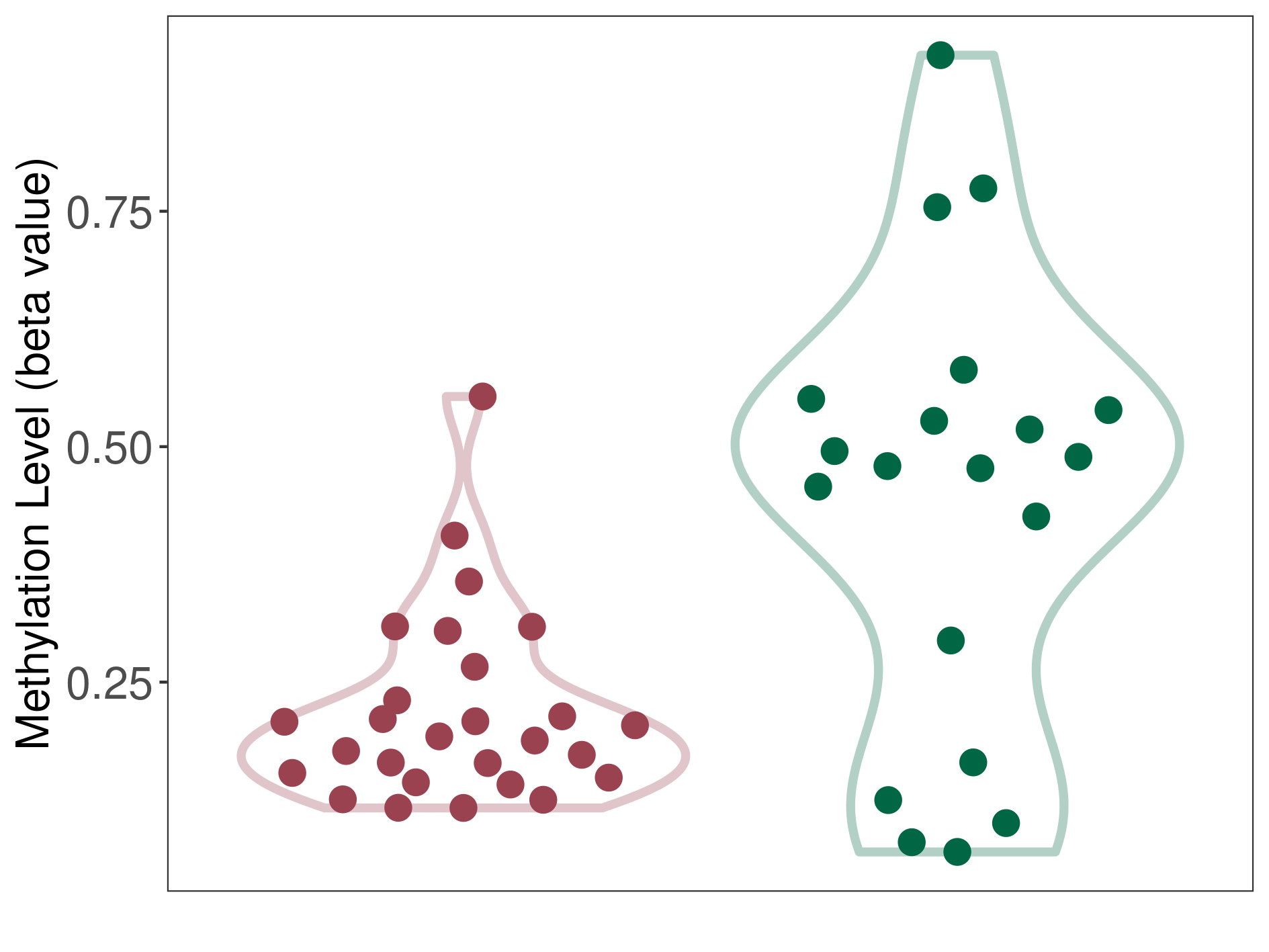
Hemangioblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC17A9 in hemangioblastoma than that in healthy individual | ||||
Studied Phenotype |
Hemangioblastoma [ICD-11:2F7C] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.000650138; Fold-change:-0.291900638; Z-score:-1.224212538 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
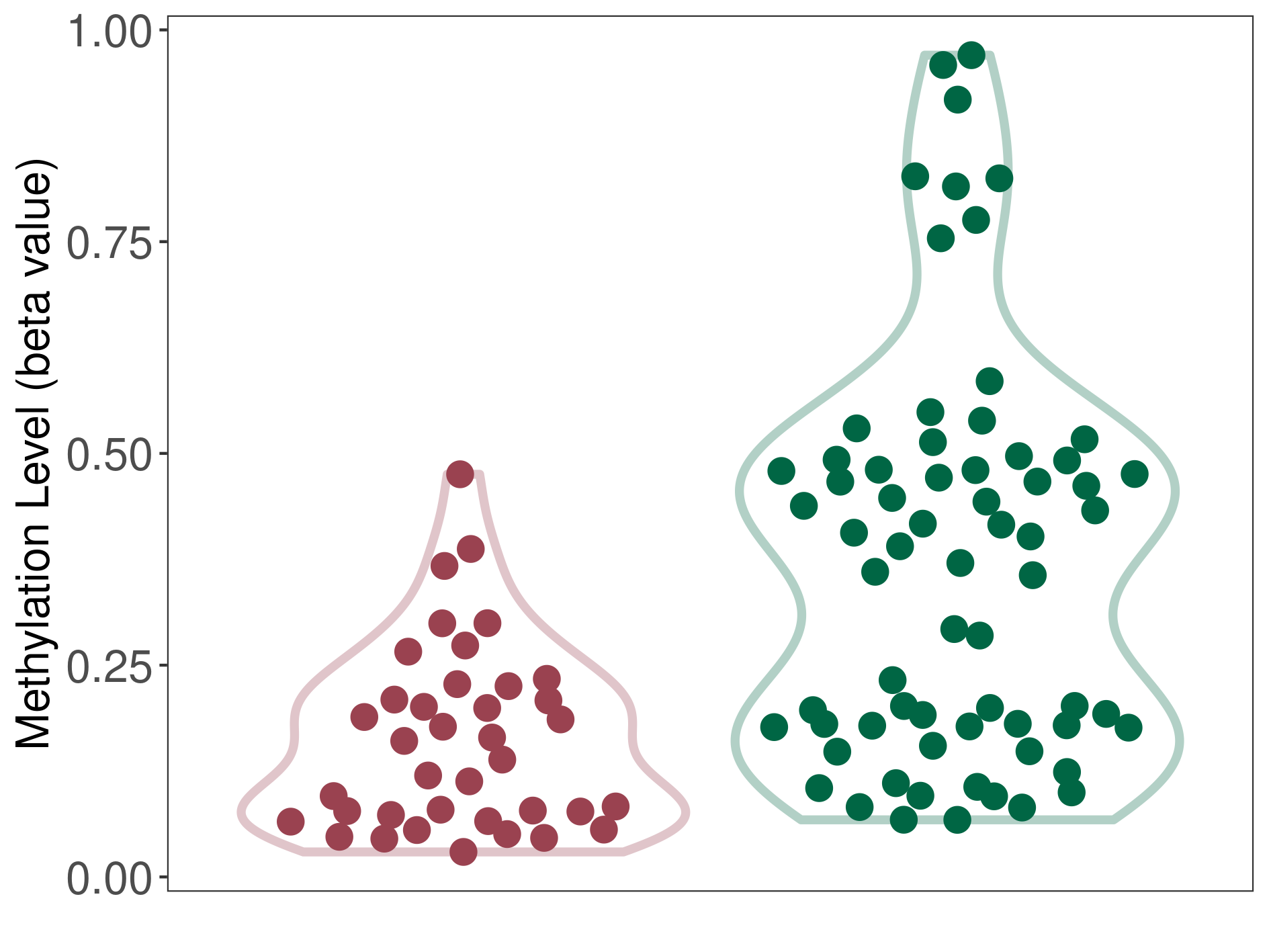
Lymphoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC17A9 in lymphoma than that in healthy individual | ||||
Studied Phenotype |
Lymphoma [ICD-11:2B30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.25E-08; Fold-change:-0.240839604; Z-score:-1.014835171 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
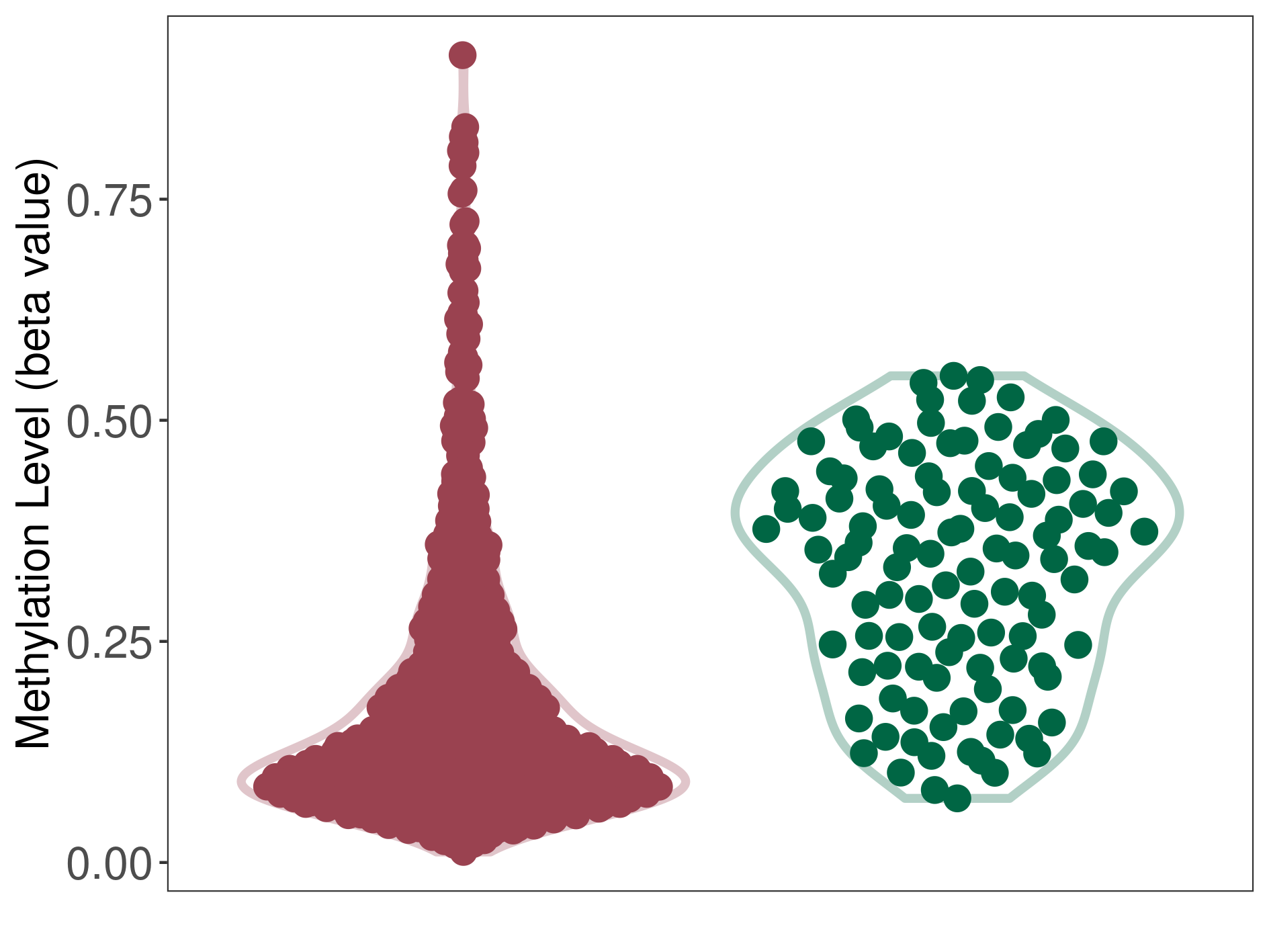
Medulloblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC17A9 in medulloblastoma than that in healthy individual | ||||
Studied Phenotype |
Medulloblastoma [ICD-11:2A00.10] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:2.17E-22; Fold-change:-0.222522677; Z-score:-1.75509628 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
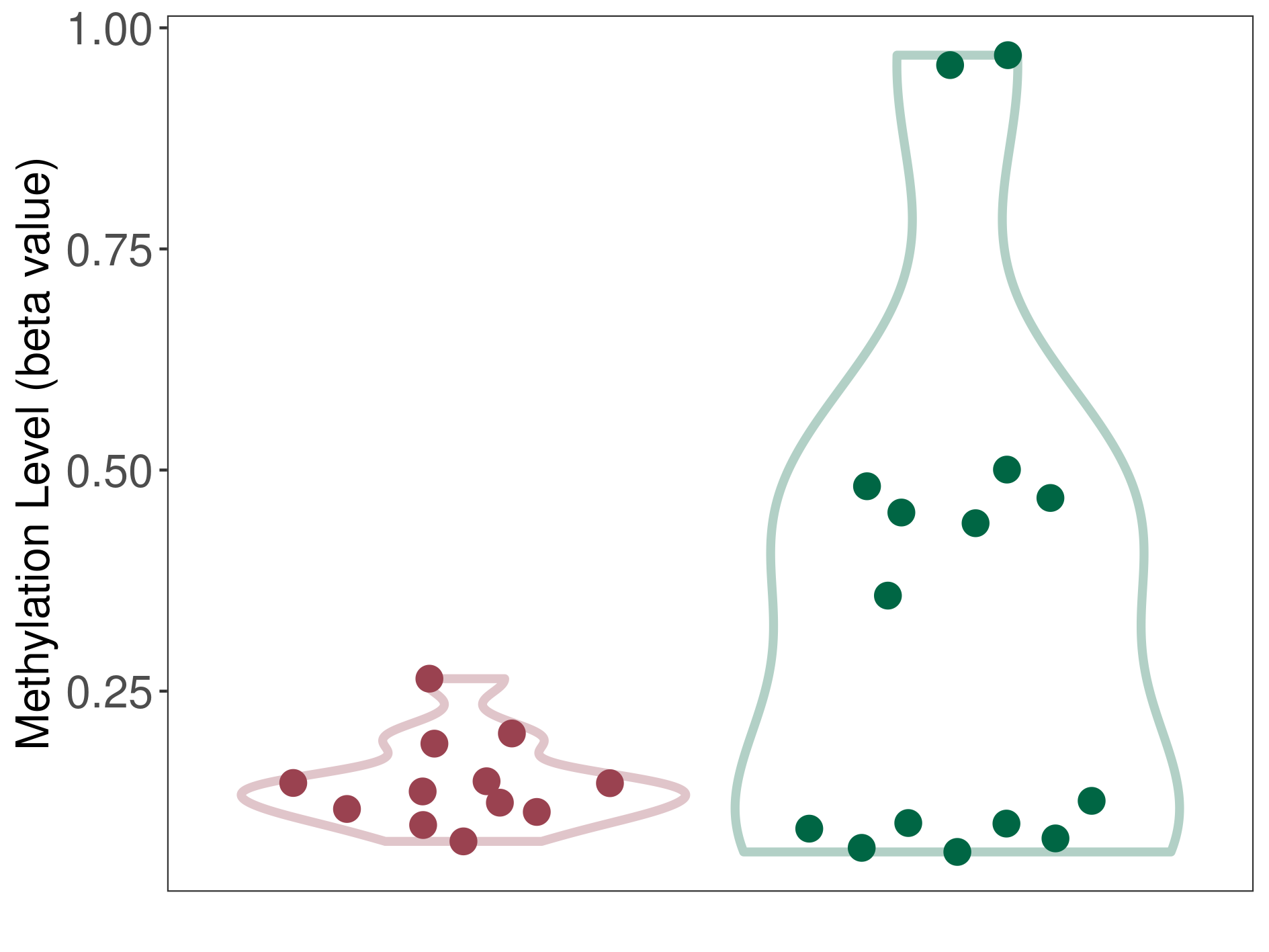
Third ventricle chordoid glioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC17A9 in third ventricle chordoid glioma than that in healthy individual | ||||
Studied Phenotype |
Third ventricle chordoid glioma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.021753412; Fold-change:-0.216754726; Z-score:-0.713374711 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
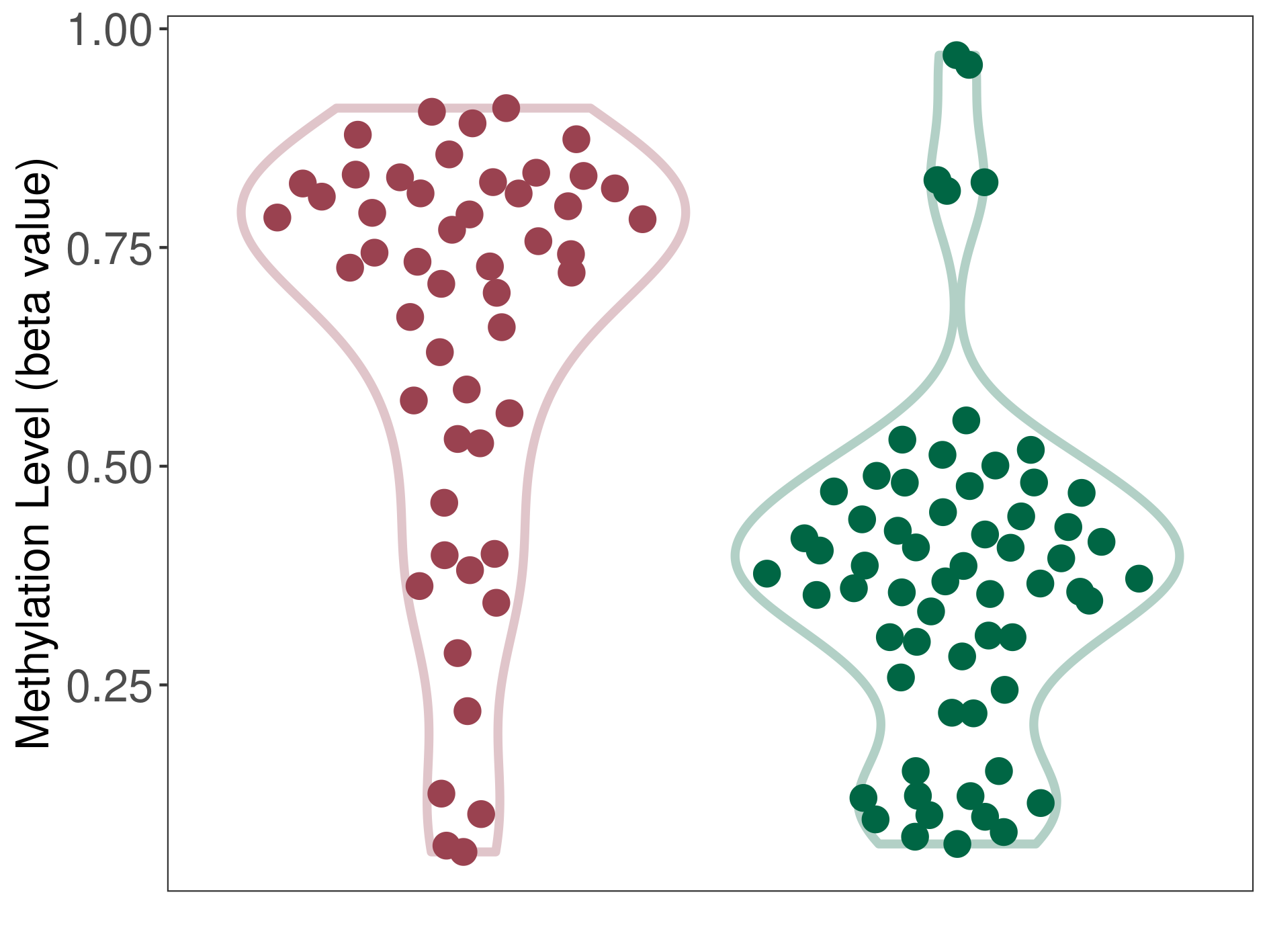
Brain neuroepithelial tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC17A9 in brain neuroepithelial tumour than that in healthy individual | ||||
Studied Phenotype |
Brain neuroepithelial tumour [ICD-11:2A00.2Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:8.24E-09; Fold-change:0.35935508; Z-score:1.786450057 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Diffuse midline glioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC17A9 in diffuse midline glioma than that in healthy individual | ||||
Studied Phenotype |
Diffuse midline glioma [ICD-11:2A00.0Z] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:2.17E-32; Fold-change:0.304267293; Z-score:1.57929554 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
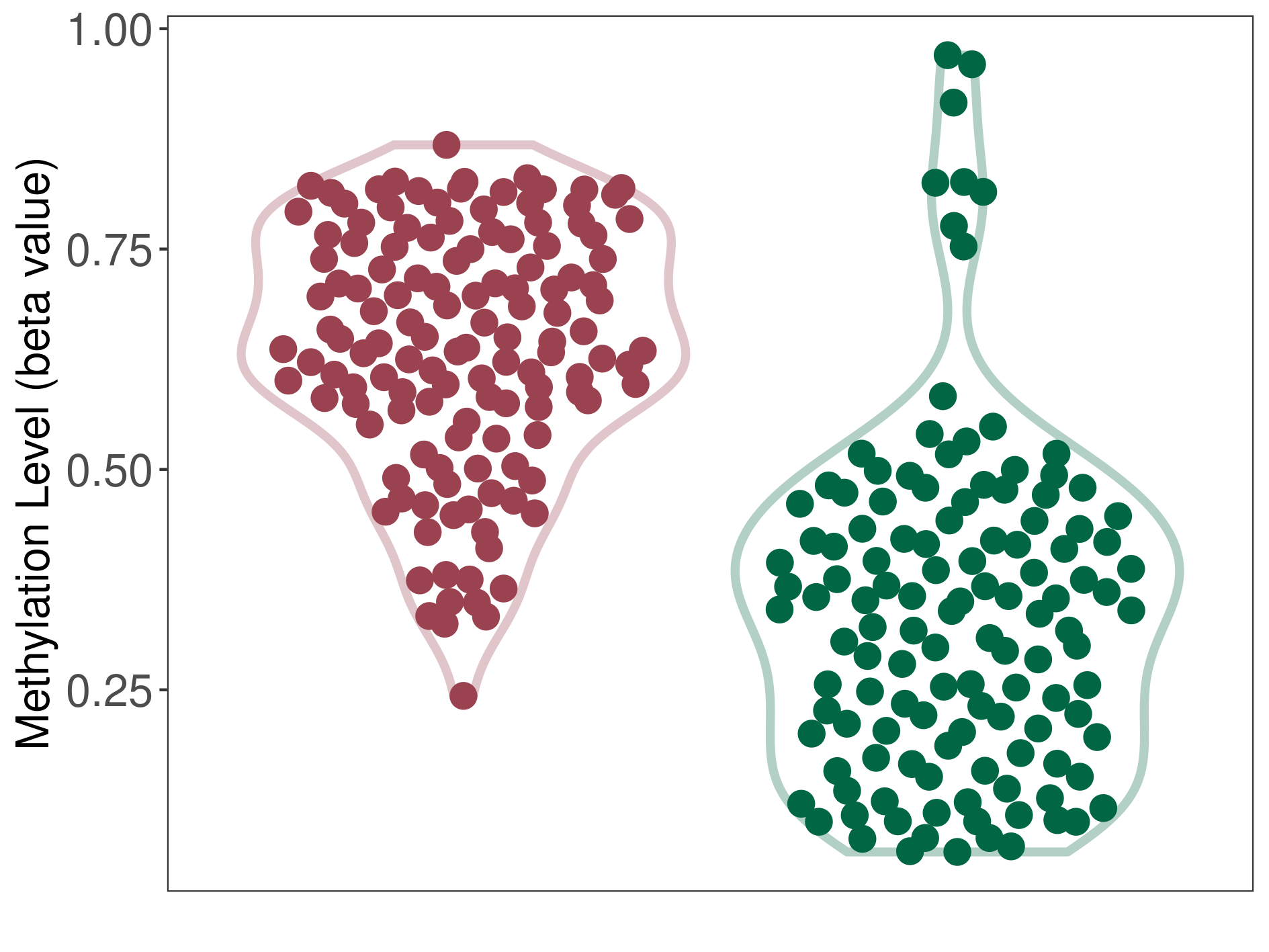
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Malignant astrocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC17A9 in malignant astrocytoma than that in healthy individual | ||||
Studied Phenotype |
Malignant astrocytoma [ICD-11:2A00.12] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:4.04E-30; Fold-change:0.362533502; Z-score:1.816657369 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
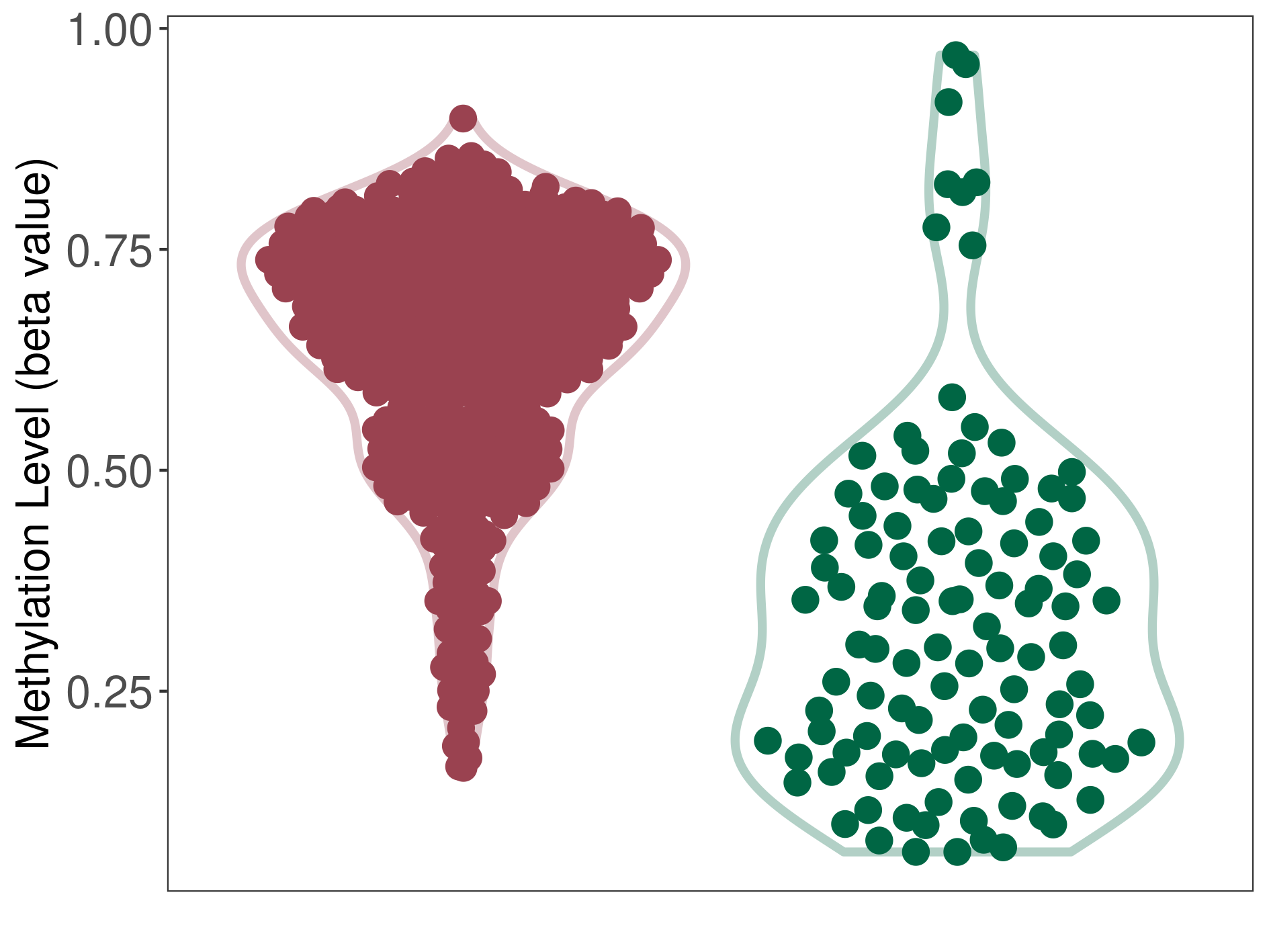
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Multilayered rosettes embryonal tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC17A9 in multilayered rosettes embryonal tumour than that in healthy individual | ||||
Studied Phenotype |
Multilayered rosettes embryonal tumour [ICD-11:2A00.1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:4.95E-13; Fold-change:0.381499314; Z-score:1.502312287 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
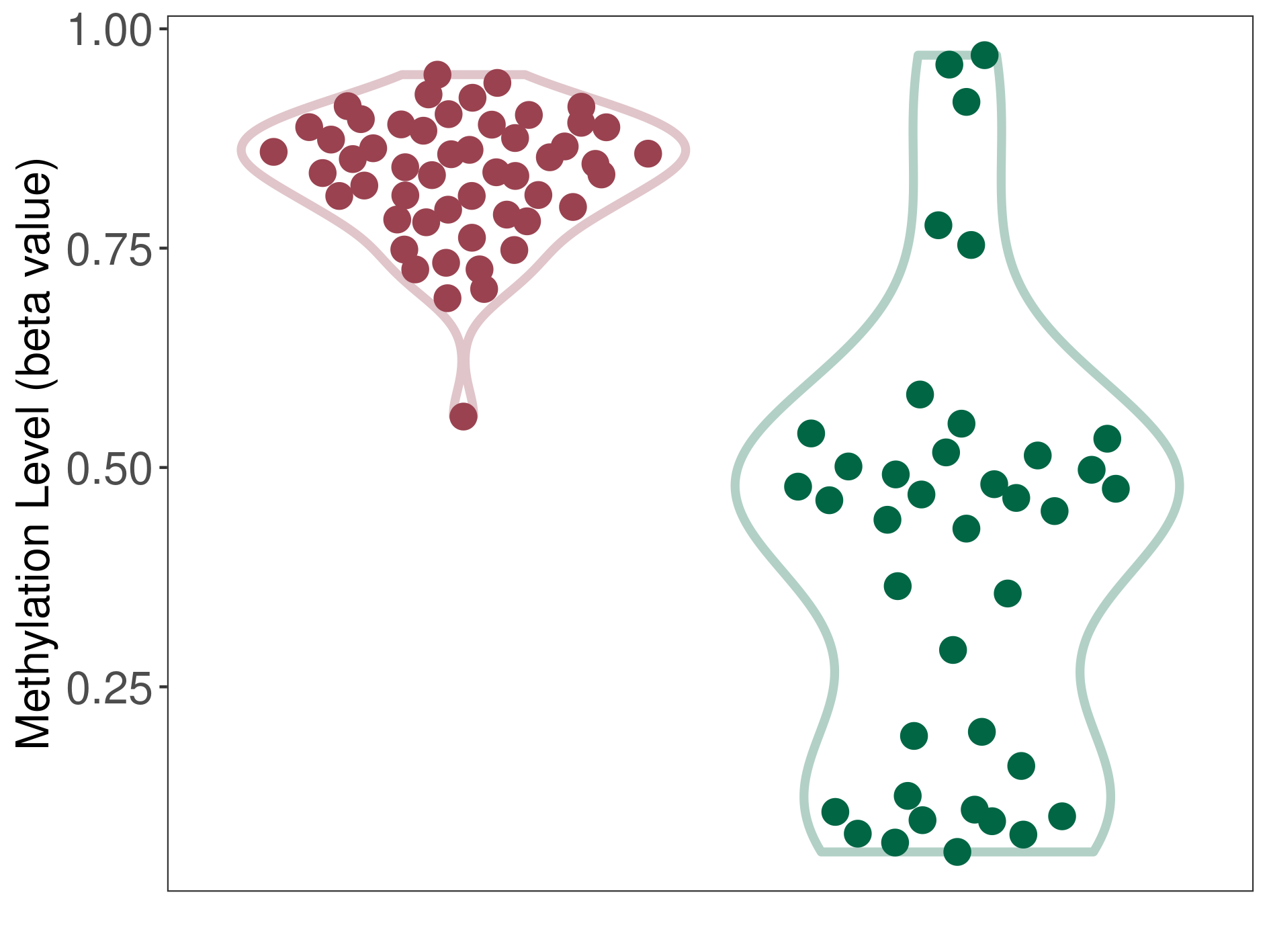
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Oligodendroglioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC17A9 in oligodendroglioma than that in healthy individual | ||||
Studied Phenotype |
Oligodendroglioma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:3.22E-19; Fold-change:0.36478069; Z-score:1.535701389 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
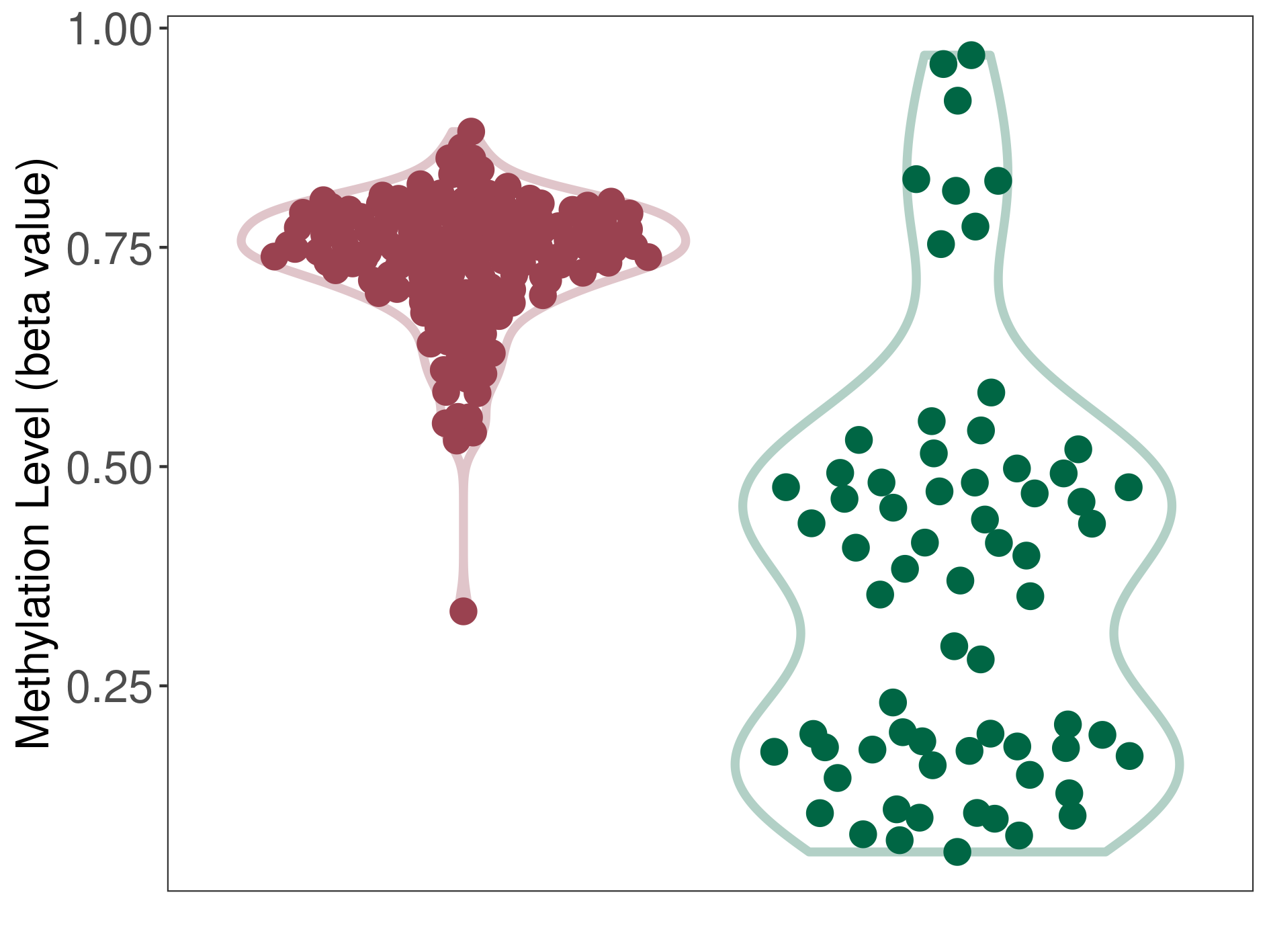
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Prostate cancer metastasis |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC17A9 in prostate cancer metastasis than that in healthy individual | ||||
Studied Phenotype |
Prostate cancer metastasis [ICD-11:2.00E+06] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:9.09E-07; Fold-change:0.429926378; Z-score:9.522657867 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
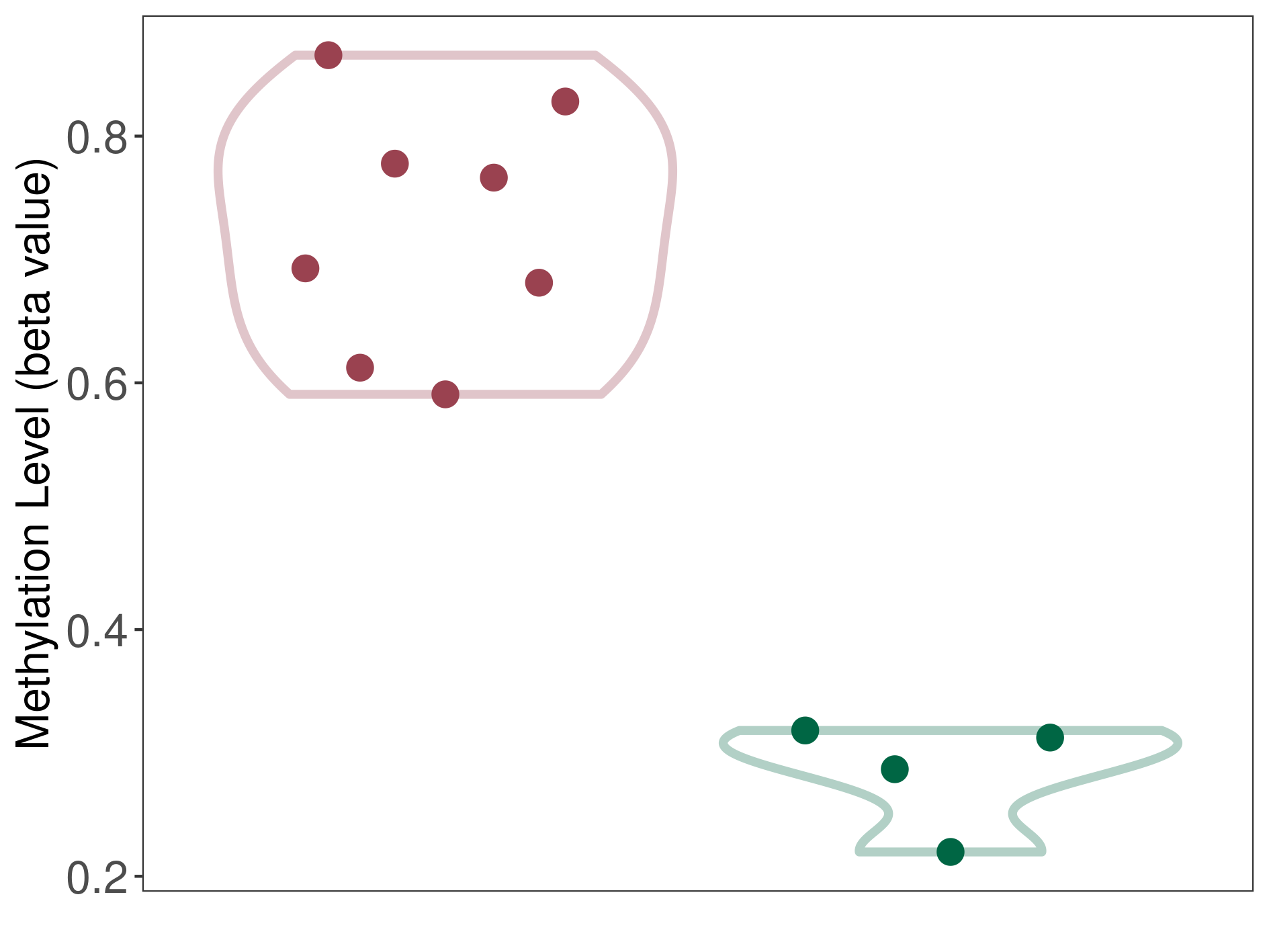
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Uterine carcinosarcoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC17A9 in uterine carcinosarcoma than that in healthy individual | ||||
Studied Phenotype |
Uterine carcinosarcoma [ICD-11:2B5F] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.001482859; Fold-change:0.303923323; Z-score:1.675513076 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
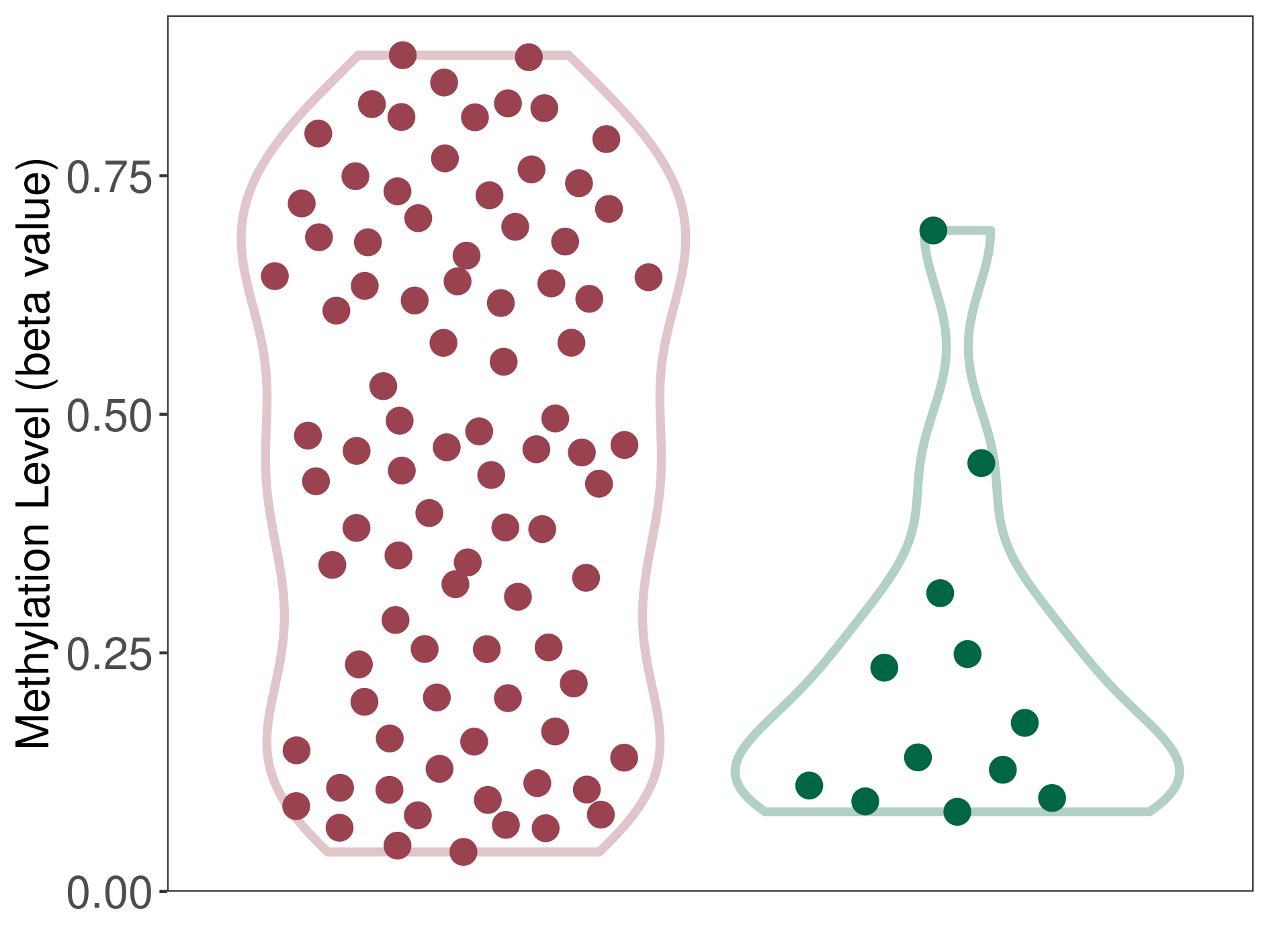
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Esthesioneuroblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC17A9 in esthesioneuroblastoma than that in healthy individual | ||||
Studied Phenotype |
Esthesioneuroblastoma [ICD-11:2D50.1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.41E-07; Fold-change:-0.406146823; Z-score:-1.500463925 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
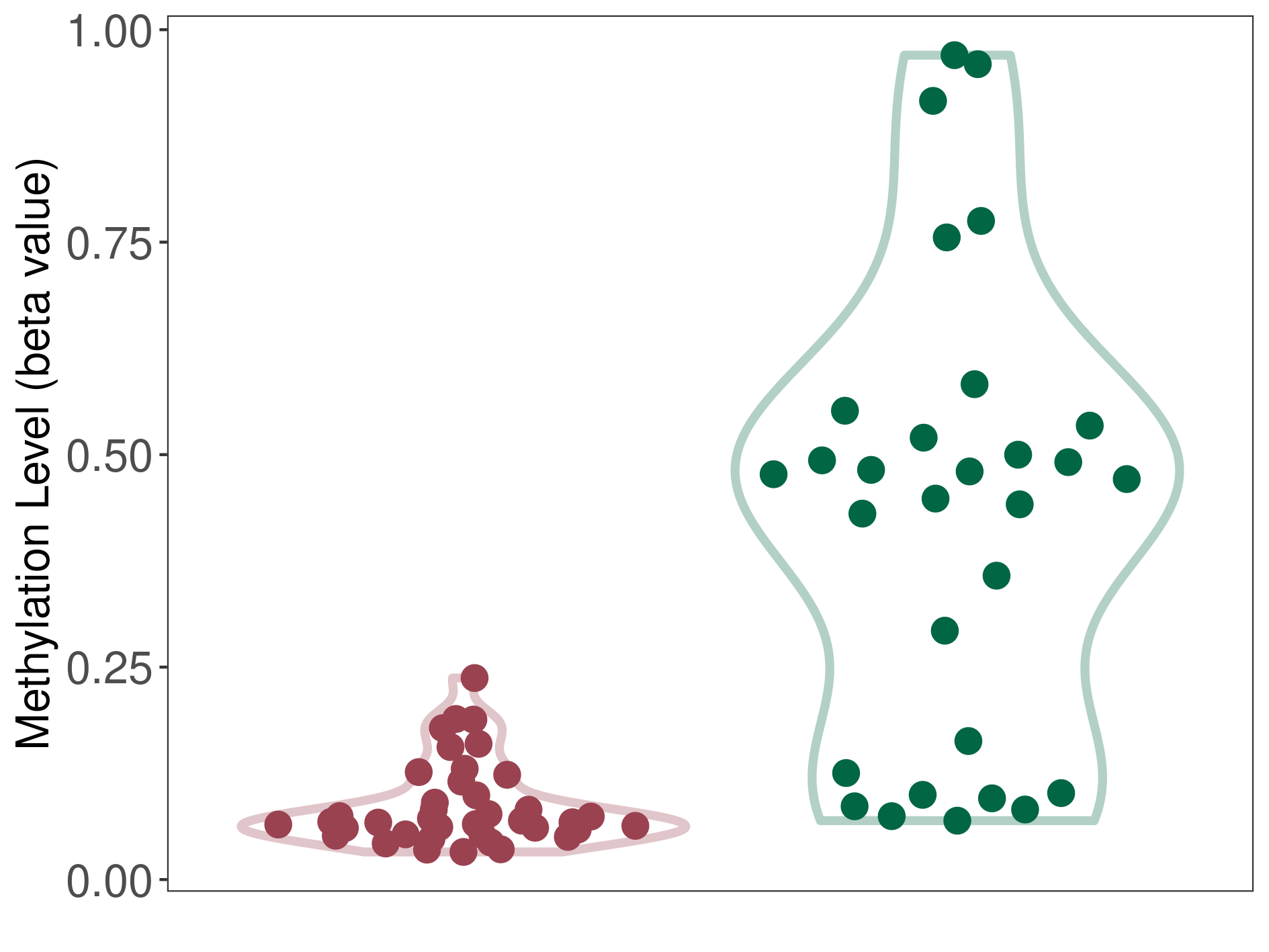
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
