Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0105 Transporter Info | ||||
| Gene Name | SLC16A14 | ||||
| Transporter Name | Monocarboxylate transporter 14 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Bladder cancer |
11 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC16A14 in bladder cancer | [ 1 ] | |||
|
Location |
5'UTR (cg24704015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:4.93E-03; Z-score:3.49E+00 | ||
|
Methylation in Case |
3.21E-01 (Median) | Methylation in Control | 2.97E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC16A14 in bladder cancer | [ 1 ] | |||
|
Location |
TSS1500 (cg02637654) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:9.64E-07; Z-score:-4.96E+00 | ||
|
Methylation in Case |
5.07E-01 (Median) | Methylation in Control | 6.87E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC16A14 in bladder cancer | [ 1 ] | |||
|
Location |
TSS1500 (cg25621735) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:2.26E-03; Z-score:-1.64E+00 | ||
|
Methylation in Case |
6.03E-01 (Median) | Methylation in Control | 6.63E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC16A14 in bladder cancer | [ 1 ] | |||
|
Location |
TSS1500 (cg20822693) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:8.75E-03; Z-score:-1.99E+00 | ||
|
Methylation in Case |
7.14E-01 (Median) | Methylation in Control | 7.89E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC16A14 in bladder cancer | [ 1 ] | |||
|
Location |
TSS200 (cg01172899) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.57E+00 | Statistic Test | p-value:2.22E-04; Z-score:-2.90E+00 | ||
|
Methylation in Case |
1.58E-01 (Median) | Methylation in Control | 2.48E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC16A14 in bladder cancer | [ 1 ] | |||
|
Location |
TSS200 (cg07490650) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:1.32E-02; Z-score:-1.24E+00 | ||
|
Methylation in Case |
4.66E-02 (Median) | Methylation in Control | 5.67E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC16A14 in bladder cancer | [ 1 ] | |||
|
Location |
Body (cg01021245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:2.19E-04; Z-score:-3.77E+00 | ||
|
Methylation in Case |
7.59E-01 (Median) | Methylation in Control | 8.52E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC16A14 in bladder cancer | [ 1 ] | |||
|
Location |
Body (cg00542545) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:5.95E-04; Z-score:-5.06E+00 | ||
|
Methylation in Case |
9.48E-01 (Median) | Methylation in Control | 9.78E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC16A14 in bladder cancer | [ 1 ] | |||
|
Location |
Body (cg21540962) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.48E-02; Z-score:-2.19E+00 | ||
|
Methylation in Case |
8.84E-01 (Median) | Methylation in Control | 9.05E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC16A14 in bladder cancer | [ 1 ] | |||
|
Location |
Body (cg20728704) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.49E-02; Z-score:-1.52E+00 | ||
|
Methylation in Case |
8.89E-01 (Median) | Methylation in Control | 9.14E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC16A14 in bladder cancer | [ 1 ] | |||
|
Location |
3'UTR (cg10277956) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-3.09E+00 | Statistic Test | p-value:2.49E-12; Z-score:-2.38E+01 | ||
|
Methylation in Case |
2.69E-01 (Median) | Methylation in Control | 8.32E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC16A14 in clear cell renal cell carcinoma | [ 2 ] | |||
|
Location |
5'UTR (cg22945872) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:2.01E-02; Z-score:5.53E-01 | ||
|
Methylation in Case |
5.41E-02 (Median) | Methylation in Control | 5.01E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC16A14 in clear cell renal cell carcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg02637654) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:1.29E-09; Z-score:-2.55E+00 | ||
|
Methylation in Case |
7.87E-01 (Median) | Methylation in Control | 8.55E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC16A14 in clear cell renal cell carcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg01172899) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.53E+00 | Statistic Test | p-value:1.45E-05; Z-score:2.38E+00 | ||
|
Methylation in Case |
3.73E-01 (Median) | Methylation in Control | 2.44E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC16A14 in clear cell renal cell carcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg13062396) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.34E+00 | Statistic Test | p-value:1.77E-04; Z-score:2.89E+00 | ||
|
Methylation in Case |
6.40E-02 (Median) | Methylation in Control | 2.74E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC16A14 in clear cell renal cell carcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg07490650) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.47E+00 | Statistic Test | p-value:2.21E-02; Z-score:1.73E+00 | ||
|
Methylation in Case |
2.17E-02 (Median) | Methylation in Control | 1.48E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colon cancer |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC16A14 in colon adenocarcinoma | [ 3 ] | |||
|
Location |
5'UTR (cg07466705) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.36E+00 | Statistic Test | p-value:7.28E-08; Z-score:2.03E+00 | ||
|
Methylation in Case |
5.31E-01 (Median) | Methylation in Control | 3.90E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC16A14 in colon adenocarcinoma | [ 3 ] | |||
|
Location |
TSS1500 (cg01178971) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.27E+00 | Statistic Test | p-value:3.28E-03; Z-score:-1.65E+00 | ||
|
Methylation in Case |
3.72E-01 (Median) | Methylation in Control | 4.70E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC16A14 in colon adenocarcinoma | [ 3 ] | |||
|
Location |
TSS200 (cg10212705) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:7.32E+00 | Statistic Test | p-value:5.03E-05; Z-score:1.84E+01 | ||
|
Methylation in Case |
3.37E-01 (Median) | Methylation in Control | 4.61E-02 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC16A14 in colon adenocarcinoma | [ 3 ] | |||
|
Location |
Body (cg00437173) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:9.44E-06; Z-score:-2.70E+00 | ||
|
Methylation in Case |
5.10E-01 (Median) | Methylation in Control | 6.01E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC16A14 in colon adenocarcinoma | [ 3 ] | |||
|
Location |
Body (cg19156231) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:2.96E-03; Z-score:1.11E+00 | ||
|
Methylation in Case |
5.54E-01 (Median) | Methylation in Control | 4.83E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
13 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC16A14 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg22945872) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:5.29E-05; Z-score:7.57E-01 | ||
|
Methylation in Case |
1.25E-01 (Median) | Methylation in Control | 1.11E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC16A14 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg24704015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:4.43E-04; Z-score:8.10E-01 | ||
|
Methylation in Case |
4.38E-01 (Median) | Methylation in Control | 4.08E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC16A14 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg26338195) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:6.06E-03; Z-score:7.01E-02 | ||
|
Methylation in Case |
2.79E-02 (Median) | Methylation in Control | 2.72E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC16A14 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg02821501) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:1.07E-02; Z-score:1.70E-01 | ||
|
Methylation in Case |
2.37E-02 (Median) | Methylation in Control | 2.28E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC16A14 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg00455164) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.09E-02; Z-score:-1.06E-01 | ||
|
Methylation in Case |
1.69E-02 (Median) | Methylation in Control | 1.78E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC16A14 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg20822693) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:9.22E-05; Z-score:-1.01E+00 | ||
|
Methylation in Case |
7.91E-01 (Median) | Methylation in Control | 8.51E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC16A14 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg02637654) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:2.89E-03; Z-score:-7.85E-01 | ||
|
Methylation in Case |
8.03E-01 (Median) | Methylation in Control | 8.33E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC16A14 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg25621735) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.86E-02; Z-score:-8.14E-01 | ||
|
Methylation in Case |
8.00E-01 (Median) | Methylation in Control | 8.30E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC16A14 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS200 (cg07490650) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:2.27E-02; Z-score:1.56E-01 | ||
|
Methylation in Case |
9.15E-02 (Median) | Methylation in Control | 8.97E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC16A14 in colorectal cancer | [ 4 ] | |||
|
Location |
Body (cg01021245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.64E-04; Z-score:-1.33E+00 | ||
|
Methylation in Case |
9.21E-01 (Median) | Methylation in Control | 9.35E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC16A14 in colorectal cancer | [ 4 ] | |||
|
Location |
Body (cg00542545) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.89E-02; Z-score:-7.66E-01 | ||
|
Methylation in Case |
9.78E-01 (Median) | Methylation in Control | 9.81E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC16A14 in colorectal cancer | [ 4 ] | |||
|
Location |
3'UTR (cg10277956) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:5.51E-09; Z-score:-2.78E+00 | ||
|
Methylation in Case |
8.28E-01 (Median) | Methylation in Control | 9.00E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
18 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
5'UTR (cg22945872) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:1.45E-06; Z-score:1.12E+00 | ||
|
Methylation in Case |
1.11E-01 (Median) | Methylation in Control | 9.21E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
5'UTR (cg26338195) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:1.09E-04; Z-score:2.91E-01 | ||
|
Methylation in Case |
3.49E-02 (Median) | Methylation in Control | 3.11E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
5'UTR (cg00483994) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:7.76E-04; Z-score:-2.81E-01 | ||
|
Methylation in Case |
8.77E-01 (Median) | Methylation in Control | 8.90E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
5'UTR (cg02821501) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:2.15E-03; Z-score:-6.09E-03 | ||
|
Methylation in Case |
3.60E-02 (Median) | Methylation in Control | 3.60E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
5'UTR (cg00455164) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:3.63E-03; Z-score:1.30E-01 | ||
|
Methylation in Case |
2.77E-02 (Median) | Methylation in Control | 2.59E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg09067747) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:1.13E-12; Z-score:-1.99E+00 | ||
|
Methylation in Case |
5.41E-01 (Median) | Methylation in Control | 6.03E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg25621735) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.99E-04; Z-score:-1.19E+00 | ||
|
Methylation in Case |
7.93E-01 (Median) | Methylation in Control | 8.20E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg20822693) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:2.06E-04; Z-score:-6.51E-01 | ||
|
Methylation in Case |
7.09E-01 (Median) | Methylation in Control | 7.55E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg02637654) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.46E-04; Z-score:-1.75E-01 | ||
|
Methylation in Case |
6.83E-01 (Median) | Methylation in Control | 6.93E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg07490650) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.58E+00 | Statistic Test | p-value:7.09E-05; Z-score:6.67E-01 | ||
|
Methylation in Case |
1.57E-01 (Median) | Methylation in Control | 9.95E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg13062396) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:8.83E-04; Z-score:4.09E-01 | ||
|
Methylation in Case |
2.25E-01 (Median) | Methylation in Control | 1.87E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg01172899) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:9.67E-03; Z-score:9.02E-01 | ||
|
Methylation in Case |
4.24E-01 (Median) | Methylation in Control | 3.52E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
1stExon (cg25177139) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.47E+00 | Statistic Test | p-value:1.06E-17; Z-score:-7.76E+00 | ||
|
Methylation in Case |
5.89E-01 (Median) | Methylation in Control | 8.66E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg16605633) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.48E+00 | Statistic Test | p-value:5.26E-16; Z-score:-2.67E+00 | ||
|
Methylation in Case |
4.96E-01 (Median) | Methylation in Control | 7.34E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg21540962) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:9.10E-06; Z-score:-1.59E+00 | ||
|
Methylation in Case |
8.50E-01 (Median) | Methylation in Control | 8.78E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg00542545) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.02E-04; Z-score:-8.91E-01 | ||
|
Methylation in Case |
9.68E-01 (Median) | Methylation in Control | 9.76E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg20728704) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:4.35E-04; Z-score:-4.18E-01 | ||
|
Methylation in Case |
7.67E-01 (Median) | Methylation in Control | 8.20E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC16A14 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
3'UTR (cg10277956) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:1.11E-07; Z-score:-1.60E+00 | ||
|
Methylation in Case |
6.74E-01 (Median) | Methylation in Control | 7.56E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Pancretic ductal adenocarcinoma |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC16A14 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
5'UTR (cg04783530) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:4.55E-02; Z-score:-1.10E-02 | ||
|
Methylation in Case |
9.15E-01 (Median) | Methylation in Control | 9.15E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC16A14 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg24168914) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:1.20E-02; Z-score:2.65E-02 | ||
|
Methylation in Case |
7.95E-02 (Median) | Methylation in Control | 7.84E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC16A14 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg05950509) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:2.66E-02; Z-score:-6.20E-01 | ||
|
Methylation in Case |
7.32E-01 (Median) | Methylation in Control | 7.74E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC16A14 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
TSS200 (cg12307787) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.19E+00 | Statistic Test | p-value:9.53E-21; Z-score:3.47E+00 | ||
|
Methylation in Case |
2.56E-01 (Median) | Methylation in Control | 1.17E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC16A14 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg05686323) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:5.43E-03; Z-score:-8.57E-01 | ||
|
Methylation in Case |
7.00E-01 (Median) | Methylation in Control | 7.82E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC16A14 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg22022988) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:4.18E-02; Z-score:6.57E-01 | ||
|
Methylation in Case |
8.95E-01 (Median) | Methylation in Control | 8.77E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC16A14 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
3'UTR (cg11105927) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:9.01E-04; Z-score:6.79E-01 | ||
|
Methylation in Case |
7.37E-01 (Median) | Methylation in Control | 7.18E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC16A14 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
5'UTR (cg26338195) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:3.75E-02; Z-score:7.24E-01 | ||
|
Methylation in Case |
8.02E-02 (Median) | Methylation in Control | 7.42E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC16A14 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg02637654) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:6.86E-03; Z-score:-3.41E-01 | ||
|
Methylation in Case |
7.59E-01 (Median) | Methylation in Control | 7.72E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC16A14 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
TSS200 (cg13062396) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:1.72E-03; Z-score:-3.81E-01 | ||
|
Methylation in Case |
8.49E-02 (Median) | Methylation in Control | 9.89E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC16A14 in papillary thyroid cancer | [ 7 ] | |||
|
Location |
Body (cg00542545) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:2.94E-02; Z-score:-4.09E-01 | ||
|
Methylation in Case |
9.49E-01 (Median) | Methylation in Control | 9.52E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC16A14 in breast cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg02637654) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:1.16E-13; Z-score:-2.12E+00 | ||
|
Methylation in Case |
6.34E-01 (Median) | Methylation in Control | 7.18E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC16A14 in breast cancer | [ 8 ] | |||
|
Location |
TSS200 (cg07490650) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.24E+00 | Statistic Test | p-value:5.06E-03; Z-score:-6.80E-01 | ||
|
Methylation in Case |
5.89E-02 (Median) | Methylation in Control | 7.30E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC16A14 in breast cancer | [ 8 ] | |||
|
Location |
TSS200 (cg01172899) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.93E-02; Z-score:-4.15E-01 | ||
|
Methylation in Case |
3.26E-01 (Median) | Methylation in Control | 3.44E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC16A14 in breast cancer | [ 8 ] | |||
|
Location |
Body (cg01021245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.83E-02; Z-score:-4.83E-01 | ||
|
Methylation in Case |
8.36E-01 (Median) | Methylation in Control | 8.48E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC16A14 in breast cancer | [ 8 ] | |||
|
Location |
3'UTR (cg10277956) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:2.07E-07; Z-score:-9.95E-01 | ||
|
Methylation in Case |
7.99E-01 (Median) | Methylation in Control | 8.44E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC16A14 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
TSS1500 (cg02637654) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:7.24E-04; Z-score:-3.15E+00 | ||
|
Methylation in Case |
6.52E-01 (Median) | Methylation in Control | 7.44E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC16A14 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
TSS1500 (cg20822693) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:8.62E-04; Z-score:-3.00E+00 | ||
|
Methylation in Case |
7.10E-01 (Median) | Methylation in Control | 8.19E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC16A14 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
TSS200 (cg07490650) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.25E+00 | Statistic Test | p-value:7.75E-03; Z-score:2.44E+00 | ||
|
Methylation in Case |
1.02E-01 (Median) | Methylation in Control | 8.16E-02 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC16A14 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
TSS200 (cg01172899) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.24E+00 | Statistic Test | p-value:2.15E-02; Z-score:1.75E+00 | ||
|
Methylation in Case |
4.01E-01 (Median) | Methylation in Control | 3.24E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC16A14 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg00542545) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.81E-03; Z-score:-1.78E+00 | ||
|
Methylation in Case |
9.64E-01 (Median) | Methylation in Control | 9.73E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC16A14 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
3'UTR (cg10277956) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:3.21E-05; Z-score:2.85E+00 | ||
|
Methylation in Case |
8.27E-01 (Median) | Methylation in Control | 7.10E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC16A14 in panic disorder | [ 10 ] | |||
|
Location |
TSS1500 (cg20822693) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:1.32E-03; Z-score:6.11E-01 | ||
|
Methylation in Case |
2.32E+00 (Median) | Methylation in Control | 2.05E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC16A14 in panic disorder | [ 10 ] | |||
|
Location |
TSS200 (cg01172899) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-8.97E-01 | Statistic Test | p-value:4.33E-03; Z-score:-4.19E-01 | ||
|
Methylation in Case |
-2.53E+00 (Median) | Methylation in Control | -2.27E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC16A14 in panic disorder | [ 10 ] | |||
|
Location |
TSS200 (cg13062396) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-9.68E-01 | Statistic Test | p-value:3.42E-02; Z-score:-2.93E-01 | ||
|
Methylation in Case |
-4.10E+00 (Median) | Methylation in Control | -3.97E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC16A14 in panic disorder | [ 10 ] | |||
|
Location |
Body (cg20728704) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:3.39E-02; Z-score:-5.10E-01 | ||
|
Methylation in Case |
1.20E+00 (Median) | Methylation in Control | 1.42E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC16A14 in panic disorder | [ 10 ] | |||
|
Location |
3'UTR (cg10277956) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:3.50E+00 | Statistic Test | p-value:2.85E-02; Z-score:-4.76E-01 | ||
|
Methylation in Case |
-3.80E-02 (Median) | Methylation in Control | 1.33E-01 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC16A14 in prostate cancer | [ 11 ] | |||
|
Location |
TSS1500 (cg27150460) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.73E+00 | Statistic Test | p-value:1.11E-02; Z-score:8.30E+00 | ||
|
Methylation in Case |
5.60E-01 (Median) | Methylation in Control | 3.23E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer metastasis |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC16A14 in prostate cancer metastasis than that in healthy individual | ||||
Studied Phenotype |
Prostate cancer metastasis [ICD-11:2.00E+06] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.001153954; Fold-change:-0.260925205; Z-score:-16.68181543 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
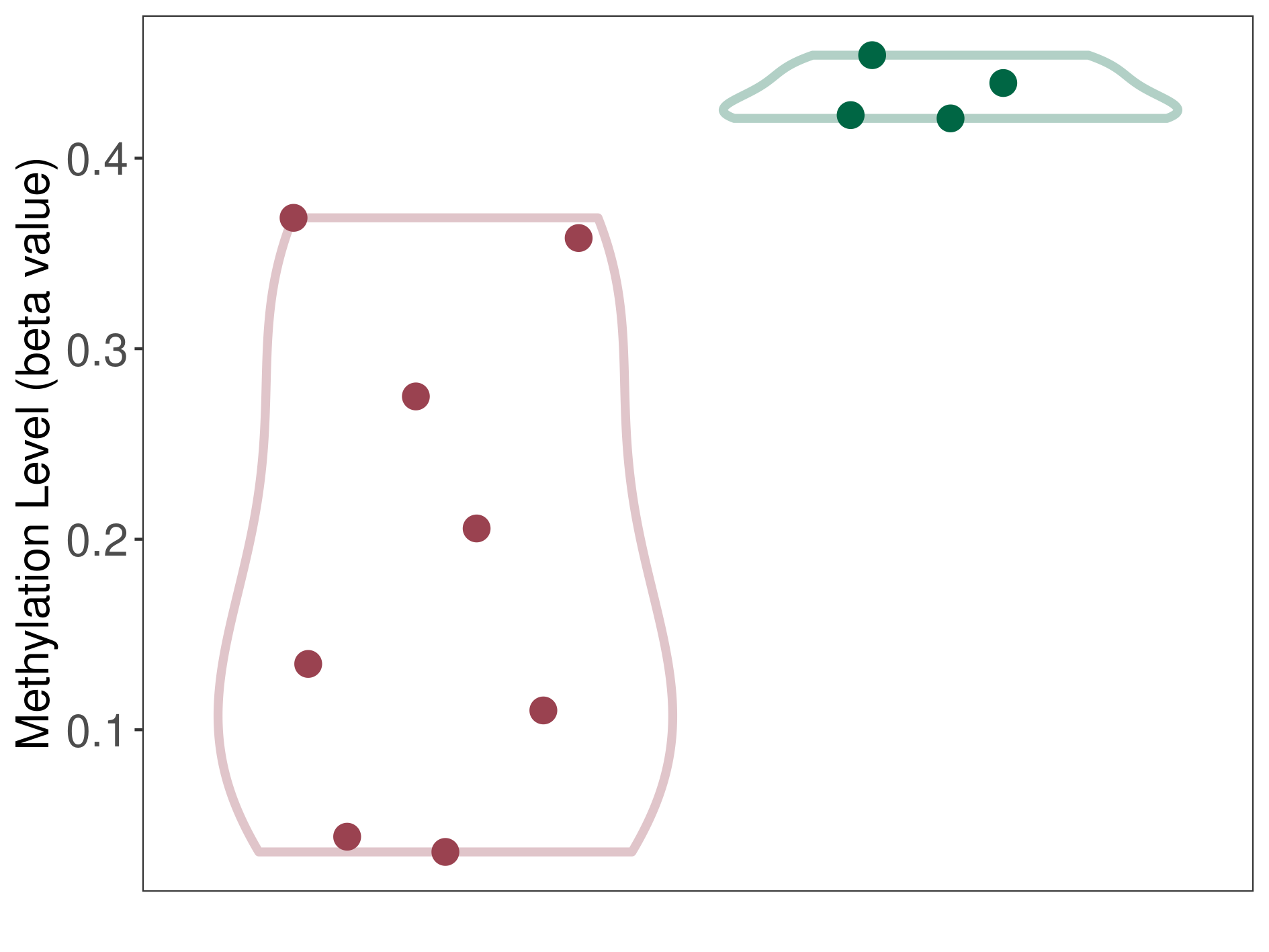
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
microRNA |
|||||
|
Unclear Phenotype |
15 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
let-7a directly targets SLC16A14 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
let-7a | miRNA Mature ID | let-7a-3p | ||
|
miRNA Sequence |
CUAUACAAUCUACUGUCUUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon2 |
let-7b directly targets SLC16A14 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
let-7b | miRNA Mature ID | let-7b-3p | ||
|
miRNA Sequence |
CUAUACAACCUACUGCCUUCCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon3 |
let-7f-1 directly targets SLC16A14 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
let-7f-1 | miRNA Mature ID | let-7f-1-3p | ||
|
miRNA Sequence |
CUAUACAAUCUAUUGCCUUCCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon4 |
miR-192 directly targets SLC16A14 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-192 | miRNA Mature ID | miR-192-5p | ||
|
miRNA Sequence |
CUGACCUAUGAAUUGACAGCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon5 |
miR-215 directly targets SLC16A14 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-215 | miRNA Mature ID | miR-215-5p | ||
|
miRNA Sequence |
AUGACCUAUGAAUUGACAGAC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon6 |
miR-300 directly targets SLC16A14 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-300 | miRNA Mature ID | miR-300 | ||
|
miRNA Sequence |
UAUACAAGGGCAGACUCUCUCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon7 |
miR-340 directly targets SLC16A14 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Sequencing | ||
|
miRNA Stemloop ID |
miR-340 | miRNA Mature ID | miR-340-5p | ||
|
miRNA Sequence |
UUAUAAAGCAAUGAGACUGAUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon8 |
miR-381 directly targets SLC16A14 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-381 | miRNA Mature ID | miR-381-3p | ||
|
miRNA Sequence |
UAUACAAGGGCAAGCUCUCUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon9 |
miR-4422 directly targets SLC16A14 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4422 | miRNA Mature ID | miR-4422 | ||
|
miRNA Sequence |
AAAAGCAUCAGGAAGUACCCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon10 |
miR-4666a directly targets SLC16A14 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4666a | miRNA Mature ID | miR-4666a-3p | ||
|
miRNA Sequence |
CAUACAAUCUGACAUGUAUUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon11 |
miR-548ad directly targets SLC16A14 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-548ad | miRNA Mature ID | miR-548ad-3p | ||
|
miRNA Sequence |
GAAAACGACAAUGACUUUUGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon12 |
miR-5589 directly targets SLC16A14 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-5589 | miRNA Mature ID | miR-5589-3p | ||
|
miRNA Sequence |
UGCACAUGGCAACCUAGCUCCCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon13 |
miR-6835 directly targets SLC16A14 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6835 | miRNA Mature ID | miR-6835-3p | ||
|
miRNA Sequence |
AAAAGCACUUUUCUGUCUCCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon14 |
miR-7849 directly targets SLC16A14 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-7849 | miRNA Mature ID | miR-7849-3p | ||
|
miRNA Sequence |
GACAAUUGUUGAUCUUGGGCCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon15 |
miR-98 directly targets SLC16A14 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-98 | miRNA Mature ID | miR-98-3p | ||
|
miRNA Sequence |
CUAUACAACUUACUACUUUCCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
