Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0088 Transporter Info | ||||
| Gene Name | SLC12A7 | ||||
| Transporter Name | Electroneutral potassium-chloride cotransporter 4 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Colon cancer |
70 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg04386206) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.64E+00 | Statistic Test | p-value:2.11E-06; Z-score:3.71E+00 | ||
|
Methylation in Case |
5.01E-01 (Median) | Methylation in Control | 3.06E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg19880462) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.45E+00 | Statistic Test | p-value:4.05E-06; Z-score:-1.51E+00 | ||
|
Methylation in Case |
1.85E-01 (Median) | Methylation in Control | 2.68E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg19300741) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.52E+00 | Statistic Test | p-value:2.32E-05; Z-score:3.57E+00 | ||
|
Methylation in Case |
3.98E-01 (Median) | Methylation in Control | 2.62E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg17258551) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.64E+00 | Statistic Test | p-value:2.79E-05; Z-score:5.40E+00 | ||
|
Methylation in Case |
3.30E-01 (Median) | Methylation in Control | 1.25E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg13952688) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.17E+00 | Statistic Test | p-value:8.70E-05; Z-score:-2.85E+00 | ||
|
Methylation in Case |
5.63E-01 (Median) | Methylation in Control | 6.61E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg16087412) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.36E+00 | Statistic Test | p-value:9.37E-05; Z-score:3.49E+00 | ||
|
Methylation in Case |
4.79E-01 (Median) | Methylation in Control | 3.53E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg00708678) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.33E+00 | Statistic Test | p-value:1.71E-04; Z-score:2.64E+00 | ||
|
Methylation in Case |
4.45E-01 (Median) | Methylation in Control | 3.34E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg08240652) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:3.68E-04; Z-score:-1.73E+00 | ||
|
Methylation in Case |
6.57E-01 (Median) | Methylation in Control | 7.55E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg13765368) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.29E+00 | Statistic Test | p-value:2.73E-03; Z-score:1.37E+00 | ||
|
Methylation in Case |
1.38E-01 (Median) | Methylation in Control | 1.07E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg00858400) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.50E+00 | Statistic Test | p-value:5.28E-09; Z-score:-6.34E+00 | ||
|
Methylation in Case |
5.27E-01 (Median) | Methylation in Control | 7.92E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg04399565) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.44E+00 | Statistic Test | p-value:2.30E-07; Z-score:-4.05E+00 | ||
|
Methylation in Case |
4.55E-01 (Median) | Methylation in Control | 6.56E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg07745624) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.66E+00 | Statistic Test | p-value:5.96E-07; Z-score:2.94E+00 | ||
|
Methylation in Case |
4.82E-01 (Median) | Methylation in Control | 2.89E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg13412615) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:3.43E-06; Z-score:-2.97E+00 | ||
|
Methylation in Case |
7.22E-01 (Median) | Methylation in Control | 8.00E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg03483626) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.37E+00 | Statistic Test | p-value:4.40E-06; Z-score:2.17E+00 | ||
|
Methylation in Case |
6.78E-01 (Median) | Methylation in Control | 4.96E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg03573068) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.76E+00 | Statistic Test | p-value:2.17E-05; Z-score:3.27E+00 | ||
|
Methylation in Case |
5.85E-01 (Median) | Methylation in Control | 3.33E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg15659974) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.26E+00 | Statistic Test | p-value:2.98E-05; Z-score:-1.75E+00 | ||
|
Methylation in Case |
4.14E-01 (Median) | Methylation in Control | 5.23E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg04676627) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.42E-04; Z-score:-2.42E+00 | ||
|
Methylation in Case |
7.82E-01 (Median) | Methylation in Control | 8.32E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg23072383) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:2.40E-04; Z-score:-6.96E-01 | ||
|
Methylation in Case |
1.07E-01 (Median) | Methylation in Control | 1.44E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg23113963) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.55E+00 | Statistic Test | p-value:2.51E-04; Z-score:2.10E+00 | ||
|
Methylation in Case |
2.90E-01 (Median) | Methylation in Control | 1.87E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg08955358) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:4.94E-04; Z-score:-1.90E+00 | ||
|
Methylation in Case |
5.80E-01 (Median) | Methylation in Control | 6.87E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg17679621) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:5.96E-04; Z-score:1.46E+00 | ||
|
Methylation in Case |
4.18E-01 (Median) | Methylation in Control | 3.42E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg21442626) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:1.18E-03; Z-score:-1.39E+00 | ||
|
Methylation in Case |
5.00E-01 (Median) | Methylation in Control | 5.80E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg23502298) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.26E+00 | Statistic Test | p-value:1.69E-03; Z-score:-1.23E+00 | ||
|
Methylation in Case |
2.21E-01 (Median) | Methylation in Control | 2.78E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg10970251) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:2.73E-03; Z-score:1.23E+00 | ||
|
Methylation in Case |
7.45E-02 (Median) | Methylation in Control | 6.53E-02 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS1500 (cg09454187) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:2.91E-03; Z-score:-2.29E+00 | ||
|
Methylation in Case |
8.00E-01 (Median) | Methylation in Control | 8.34E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg24607642) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:8.57E-09; Z-score:-2.28E+00 | ||
|
Methylation in Case |
3.96E-01 (Median) | Methylation in Control | 5.40E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg07808555) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.64E+00 | Statistic Test | p-value:1.71E-08; Z-score:2.79E+00 | ||
|
Methylation in Case |
5.37E-01 (Median) | Methylation in Control | 2.03E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg12882697) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.72E+00 | Statistic Test | p-value:7.24E-08; Z-score:5.36E+00 | ||
|
Methylation in Case |
4.81E-01 (Median) | Methylation in Control | 1.77E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg02584459) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:3.90E+00 | Statistic Test | p-value:1.02E-07; Z-score:6.45E+00 | ||
|
Methylation in Case |
5.40E-01 (Median) | Methylation in Control | 1.38E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg14588399) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.47E+00 | Statistic Test | p-value:2.02E-06; Z-score:-2.07E+00 | ||
|
Methylation in Case |
3.39E-01 (Median) | Methylation in Control | 4.98E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg03738669) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.24E+00 | Statistic Test | p-value:2.39E-06; Z-score:4.00E+00 | ||
|
Methylation in Case |
6.82E-01 (Median) | Methylation in Control | 5.50E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg09439192) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:6.60E-05; Z-score:-2.20E+00 | ||
|
Methylation in Case |
6.14E-01 (Median) | Methylation in Control | 7.12E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg18723442) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.21E+00 | Statistic Test | p-value:7.40E-05; Z-score:-2.84E+00 | ||
|
Methylation in Case |
5.28E-01 (Median) | Methylation in Control | 6.40E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg00895997) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.28E+00 | Statistic Test | p-value:1.18E-04; Z-score:-1.38E+00 | ||
|
Methylation in Case |
3.37E-01 (Median) | Methylation in Control | 4.30E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg22521269) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.58E+00 | Statistic Test | p-value:3.85E-04; Z-score:2.40E+00 | ||
|
Methylation in Case |
4.12E-01 (Median) | Methylation in Control | 2.60E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg26948274) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:4.09E-04; Z-score:-1.43E+00 | ||
|
Methylation in Case |
4.29E-01 (Median) | Methylation in Control | 4.70E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon37 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg01050423) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.68E+00 | Statistic Test | p-value:3.68E-03; Z-score:-8.46E-01 | ||
|
Methylation in Case |
1.08E-01 (Median) | Methylation in Control | 1.83E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon38 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
1stExon (cg18481230) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.47E+00 | Statistic Test | p-value:5.27E-07; Z-score:7.24E+00 | ||
|
Methylation in Case |
4.02E-01 (Median) | Methylation in Control | 1.63E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon39 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
1stExon (cg15230781) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.81E+00 | Statistic Test | p-value:2.89E-05; Z-score:2.42E+00 | ||
|
Methylation in Case |
5.25E-01 (Median) | Methylation in Control | 2.91E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon40 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
1stExon (cg06987672) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.35E+00 | Statistic Test | p-value:1.18E-03; Z-score:1.52E+00 | ||
|
Methylation in Case |
2.81E-01 (Median) | Methylation in Control | 2.08E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon41 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg12451099) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.45E+00 | Statistic Test | p-value:1.60E-08; Z-score:2.61E+00 | ||
|
Methylation in Case |
6.71E-01 (Median) | Methylation in Control | 4.63E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon42 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg06943912) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.45E+00 | Statistic Test | p-value:8.55E-07; Z-score:-2.34E+00 | ||
|
Methylation in Case |
4.60E-01 (Median) | Methylation in Control | 6.65E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon43 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg19326876) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.39E+00 | Statistic Test | p-value:1.62E-06; Z-score:2.01E+00 | ||
|
Methylation in Case |
5.35E-01 (Median) | Methylation in Control | 3.84E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon44 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg00420510) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:5.89E-06; Z-score:1.71E+00 | ||
|
Methylation in Case |
7.09E-01 (Median) | Methylation in Control | 6.42E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon45 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg06943853) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:1.02E-04; Z-score:-2.74E+00 | ||
|
Methylation in Case |
7.46E-01 (Median) | Methylation in Control | 8.35E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon46 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg26478036) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.19E+00 | Statistic Test | p-value:1.02E-04; Z-score:-3.90E+00 | ||
|
Methylation in Case |
6.79E-01 (Median) | Methylation in Control | 8.05E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon47 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg17424999) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.44E+00 | Statistic Test | p-value:1.52E-04; Z-score:1.56E+00 | ||
|
Methylation in Case |
4.06E-01 (Median) | Methylation in Control | 2.82E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon48 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg25601446) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:1.72E-04; Z-score:-1.55E+00 | ||
|
Methylation in Case |
7.24E-01 (Median) | Methylation in Control | 7.79E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon49 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg00652223) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.20E+00 | Statistic Test | p-value:1.96E-04; Z-score:-1.78E+00 | ||
|
Methylation in Case |
6.56E-01 (Median) | Methylation in Control | 7.86E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon50 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg02929434) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:1.98E-04; Z-score:-3.27E+00 | ||
|
Methylation in Case |
6.38E-01 (Median) | Methylation in Control | 7.30E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon51 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg04994795) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:3.37E-04; Z-score:-1.46E+00 | ||
|
Methylation in Case |
5.88E-01 (Median) | Methylation in Control | 6.71E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon52 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg19752565) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:3.79E-04; Z-score:-2.69E+00 | ||
|
Methylation in Case |
5.49E-01 (Median) | Methylation in Control | 6.68E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon53 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg06228648) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:4.13E-04; Z-score:-2.35E+00 | ||
|
Methylation in Case |
5.95E-01 (Median) | Methylation in Control | 7.24E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon54 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg25025399) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:7.70E-04; Z-score:-7.51E-01 | ||
|
Methylation in Case |
8.43E-01 (Median) | Methylation in Control | 8.60E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon55 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg19754709) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.24E+00 | Statistic Test | p-value:9.05E-04; Z-score:-3.99E+00 | ||
|
Methylation in Case |
5.80E-01 (Median) | Methylation in Control | 7.18E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon56 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg24602704) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.16E-03; Z-score:-8.65E-01 | ||
|
Methylation in Case |
8.42E-01 (Median) | Methylation in Control | 8.70E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon57 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg07878951) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:1.29E-03; Z-score:-3.14E+00 | ||
|
Methylation in Case |
7.09E-01 (Median) | Methylation in Control | 7.72E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon58 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg17379325) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.41E+00 | Statistic Test | p-value:1.52E-03; Z-score:2.28E+00 | ||
|
Methylation in Case |
3.87E-01 (Median) | Methylation in Control | 2.74E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon59 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg24041118) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:1.73E-03; Z-score:-1.38E+00 | ||
|
Methylation in Case |
7.75E-01 (Median) | Methylation in Control | 8.29E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon60 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg14118062) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.40E+00 | Statistic Test | p-value:1.92E-03; Z-score:-9.13E-01 | ||
|
Methylation in Case |
1.19E-01 (Median) | Methylation in Control | 1.66E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon61 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg01021245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.99E-03; Z-score:-2.35E+00 | ||
|
Methylation in Case |
8.33E-01 (Median) | Methylation in Control | 8.66E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon62 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg13755144) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.06E-03; Z-score:-1.35E+00 | ||
|
Methylation in Case |
8.21E-01 (Median) | Methylation in Control | 8.63E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon63 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg02286270) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:2.35E-03; Z-score:-1.47E+00 | ||
|
Methylation in Case |
6.55E-01 (Median) | Methylation in Control | 7.22E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon64 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg02107240) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:3.23E-03; Z-score:-2.15E+00 | ||
|
Methylation in Case |
7.39E-01 (Median) | Methylation in Control | 7.81E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon65 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg19924544) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:3.44E-03; Z-score:9.47E-01 | ||
|
Methylation in Case |
7.29E-01 (Median) | Methylation in Control | 6.67E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon66 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg24924505) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:4.24E-03; Z-score:-1.30E+00 | ||
|
Methylation in Case |
3.93E-01 (Median) | Methylation in Control | 4.37E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon67 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
Body (cg02496728) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.24E+00 | Statistic Test | p-value:4.49E-03; Z-score:-1.40E+00 | ||
|
Methylation in Case |
8.61E-02 (Median) | Methylation in Control | 1.93E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon68 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
3'UTR (cg10481065) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.88E+00 | Statistic Test | p-value:1.77E-08; Z-score:3.74E+00 | ||
|
Methylation in Case |
5.87E-01 (Median) | Methylation in Control | 3.12E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon69 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
3'UTR (cg06162589) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.44E+00 | Statistic Test | p-value:1.40E-07; Z-score:-3.05E+00 | ||
|
Methylation in Case |
4.39E-01 (Median) | Methylation in Control | 6.34E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon70 |
Methylation of SLC12A7 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
3'UTR (cg22513901) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.51E+00 | Statistic Test | p-value:2.95E-03; Z-score:3.47E+00 | ||
|
Methylation in Case |
3.96E-01 (Median) | Methylation in Control | 2.63E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Pancretic ductal adenocarcinoma |
71 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
5'UTR (cg08521987) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:5.24E+00 | Statistic Test | p-value:3.90E-43; Z-score:8.18E+00 | ||
|
Methylation in Case |
4.15E-01 (Median) | Methylation in Control | 7.93E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
5'UTR (cg03746851) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.26E+00 | Statistic Test | p-value:1.81E-10; Z-score:1.95E+00 | ||
|
Methylation in Case |
6.96E-01 (Median) | Methylation in Control | 5.52E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
5'UTR (ch.20.983686F) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.27E+00 | Statistic Test | p-value:3.07E-04; Z-score:-2.90E-01 | ||
|
Methylation in Case |
6.84E-02 (Median) | Methylation in Control | 8.66E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg12778476) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.28E+00 | Statistic Test | p-value:6.83E-14; Z-score:1.22E+00 | ||
|
Methylation in Case |
1.13E-01 (Median) | Methylation in Control | 8.84E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg08317694) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.38E+00 | Statistic Test | p-value:6.05E-12; Z-score:2.30E+00 | ||
|
Methylation in Case |
6.67E-01 (Median) | Methylation in Control | 4.83E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg00543869) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:8.20E-11; Z-score:-1.20E+00 | ||
|
Methylation in Case |
7.84E-01 (Median) | Methylation in Control | 8.11E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg07915221) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:2.37E-07; Z-score:-7.77E-01 | ||
|
Methylation in Case |
8.86E-01 (Median) | Methylation in Control | 9.09E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg24282826) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.53E+00 | Statistic Test | p-value:3.92E-07; Z-score:-1.66E+00 | ||
|
Methylation in Case |
1.51E-01 (Median) | Methylation in Control | 2.31E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg03993463) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:1.11E-06; Z-score:1.22E+00 | ||
|
Methylation in Case |
8.11E-01 (Median) | Methylation in Control | 7.27E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg07139301) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.30E+00 | Statistic Test | p-value:1.26E-05; Z-score:-9.98E-01 | ||
|
Methylation in Case |
8.21E-02 (Median) | Methylation in Control | 1.89E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg06841846) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.69E+00 | Statistic Test | p-value:6.27E-05; Z-score:-9.85E-01 | ||
|
Methylation in Case |
1.09E-01 (Median) | Methylation in Control | 1.84E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg18083248) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.18E+00 | Statistic Test | p-value:8.04E-04; Z-score:8.05E-01 | ||
|
Methylation in Case |
4.61E-01 (Median) | Methylation in Control | 3.91E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg12225685) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.66E-03; Z-score:-1.59E-01 | ||
|
Methylation in Case |
8.61E-01 (Median) | Methylation in Control | 8.64E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg15099037) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:5.87E-03; Z-score:-5.87E-01 | ||
|
Methylation in Case |
5.52E-01 (Median) | Methylation in Control | 5.69E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg07827943) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.58E-02; Z-score:-1.63E-01 | ||
|
Methylation in Case |
2.20E-01 (Median) | Methylation in Control | 2.25E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg01337931) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:2.17E-02; Z-score:4.19E-01 | ||
|
Methylation in Case |
7.32E-01 (Median) | Methylation in Control | 7.04E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg03728296) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:4.28E-02; Z-score:-3.48E-01 | ||
|
Methylation in Case |
4.32E-01 (Median) | Methylation in Control | 4.57E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg10951120) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.87E+00 | Statistic Test | p-value:1.20E-20; Z-score:3.03E+00 | ||
|
Methylation in Case |
8.68E-02 (Median) | Methylation in Control | 4.63E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg16164276) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.51E+00 | Statistic Test | p-value:3.76E-15; Z-score:1.37E+00 | ||
|
Methylation in Case |
1.78E-01 (Median) | Methylation in Control | 1.18E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg21520042) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:9.07E-11; Z-score:1.32E+00 | ||
|
Methylation in Case |
9.43E-02 (Median) | Methylation in Control | 7.82E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg26511326) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:8.81E-07; Z-score:1.47E+00 | ||
|
Methylation in Case |
7.52E-01 (Median) | Methylation in Control | 7.00E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg27619475) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-3.76E+00 | Statistic Test | p-value:2.11E-06; Z-score:-1.61E+00 | ||
|
Methylation in Case |
1.05E-01 (Median) | Methylation in Control | 3.94E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg26808921) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:1.26E-04; Z-score:-5.88E-01 | ||
|
Methylation in Case |
5.94E-02 (Median) | Methylation in Control | 8.07E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg07287120) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:1.70E-03; Z-score:-7.69E-01 | ||
|
Methylation in Case |
5.75E-02 (Median) | Methylation in Control | 6.56E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg10864955) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:2.68E-03; Z-score:-5.45E-01 | ||
|
Methylation in Case |
5.80E-02 (Median) | Methylation in Control | 6.23E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg24188017) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:3.17E-03; Z-score:-5.48E-01 | ||
|
Methylation in Case |
8.46E-01 (Median) | Methylation in Control | 8.78E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg05090759) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.21E+00 | Statistic Test | p-value:3.88E-03; Z-score:-9.59E-01 | ||
|
Methylation in Case |
2.95E-01 (Median) | Methylation in Control | 3.58E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg25201783) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.20E-02; Z-score:-2.01E-01 | ||
|
Methylation in Case |
6.37E-02 (Median) | Methylation in Control | 6.71E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg05760641) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:2.43E-02; Z-score:-5.75E-01 | ||
|
Methylation in Case |
2.56E-02 (Median) | Methylation in Control | 2.93E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg00065408) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:3.96E-02; Z-score:-2.53E-01 | ||
|
Methylation in Case |
8.56E-02 (Median) | Methylation in Control | 9.10E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
1stExon (cg06089988) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.19E+00 | Statistic Test | p-value:6.99E-04; Z-score:-6.74E-01 | ||
|
Methylation in Case |
1.13E-01 (Median) | Methylation in Control | 1.34E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
1stExon (cg19196684) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:1.11E-02; Z-score:5.81E-01 | ||
|
Methylation in Case |
7.82E-01 (Median) | Methylation in Control | 7.57E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
1stExon (cg13630845) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:2.24E-02; Z-score:-6.61E-01 | ||
|
Methylation in Case |
3.68E-02 (Median) | Methylation in Control | 4.27E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg06178383) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.31E+00 | Statistic Test | p-value:8.27E-13; Z-score:-2.37E+00 | ||
|
Methylation in Case |
4.16E-01 (Median) | Methylation in Control | 5.45E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg23530917) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.99E-12; Z-score:-1.60E+00 | ||
|
Methylation in Case |
8.90E-01 (Median) | Methylation in Control | 9.10E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg04804543) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:3.57E-11; Z-score:-1.99E+00 | ||
|
Methylation in Case |
7.01E-01 (Median) | Methylation in Control | 8.01E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon37 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg11480762) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.34E+00 | Statistic Test | p-value:3.05E-09; Z-score:-1.85E+00 | ||
|
Methylation in Case |
5.32E-01 (Median) | Methylation in Control | 7.15E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon38 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg05034603) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.17E+00 | Statistic Test | p-value:4.08E-07; Z-score:-1.73E+00 | ||
|
Methylation in Case |
3.93E-01 (Median) | Methylation in Control | 4.61E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon39 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg26434400) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.42E-06; Z-score:-4.92E-01 | ||
|
Methylation in Case |
8.90E-01 (Median) | Methylation in Control | 9.00E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon40 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg01746503) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.02E-05; Z-score:-5.27E-01 | ||
|
Methylation in Case |
8.65E-01 (Median) | Methylation in Control | 8.72E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon41 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg07338584) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.38E-05; Z-score:-3.67E-01 | ||
|
Methylation in Case |
8.14E-01 (Median) | Methylation in Control | 8.25E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon42 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg13726878) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:4.69E-05; Z-score:1.49E+00 | ||
|
Methylation in Case |
7.63E-01 (Median) | Methylation in Control | 6.73E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon43 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg18666630) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:1.68E-04; Z-score:-1.07E+00 | ||
|
Methylation in Case |
4.89E-01 (Median) | Methylation in Control | 5.96E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon44 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg19323951) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-3.09E+00 | Statistic Test | p-value:1.77E-04; Z-score:-1.75E+00 | ||
|
Methylation in Case |
1.27E-01 (Median) | Methylation in Control | 3.93E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon45 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg12582965) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:2.06E-04; Z-score:9.73E-01 | ||
|
Methylation in Case |
8.67E-01 (Median) | Methylation in Control | 7.54E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon46 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg11943056) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:2.68E-04; Z-score:-8.53E-01 | ||
|
Methylation in Case |
5.84E-01 (Median) | Methylation in Control | 6.38E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon47 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg14001035) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.23E+00 | Statistic Test | p-value:3.15E-04; Z-score:1.17E+00 | ||
|
Methylation in Case |
8.10E-01 (Median) | Methylation in Control | 6.58E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon48 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg04308167) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:3.49E-04; Z-score:-9.24E-01 | ||
|
Methylation in Case |
6.39E-01 (Median) | Methylation in Control | 7.22E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon49 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg07620167) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:4.01E-04; Z-score:-4.66E-01 | ||
|
Methylation in Case |
4.47E-01 (Median) | Methylation in Control | 4.76E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon50 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg12092346) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:4.73E-04; Z-score:9.29E-01 | ||
|
Methylation in Case |
7.60E-01 (Median) | Methylation in Control | 7.04E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon51 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg13071812) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.31E+00 | Statistic Test | p-value:6.58E-04; Z-score:-1.06E+00 | ||
|
Methylation in Case |
1.86E-01 (Median) | Methylation in Control | 2.44E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon52 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg00317626) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:7.37E-04; Z-score:8.49E-01 | ||
|
Methylation in Case |
7.41E-01 (Median) | Methylation in Control | 6.90E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon53 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg08225137) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:7.72E-04; Z-score:2.41E-01 | ||
|
Methylation in Case |
8.38E-01 (Median) | Methylation in Control | 8.33E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon54 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg10956818) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:8.95E-04; Z-score:-5.61E-01 | ||
|
Methylation in Case |
8.44E-01 (Median) | Methylation in Control | 8.69E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon55 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg08448341) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:1.74E-03; Z-score:6.29E-01 | ||
|
Methylation in Case |
8.69E-01 (Median) | Methylation in Control | 8.11E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon56 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg06012269) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:1.94E-03; Z-score:1.58E-01 | ||
|
Methylation in Case |
9.00E-01 (Median) | Methylation in Control | 8.80E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon57 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg11742688) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:5.97E-03; Z-score:-3.17E-01 | ||
|
Methylation in Case |
8.83E-01 (Median) | Methylation in Control | 8.94E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon58 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg03372042) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:6.10E-03; Z-score:1.10E+00 | ||
|
Methylation in Case |
7.77E-01 (Median) | Methylation in Control | 6.39E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon59 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg13402942) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:9.02E-03; Z-score:8.13E-01 | ||
|
Methylation in Case |
6.20E-01 (Median) | Methylation in Control | 5.88E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon60 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg26445561) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:9.90E-03; Z-score:-4.03E-01 | ||
|
Methylation in Case |
8.08E-01 (Median) | Methylation in Control | 8.25E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon61 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg03807914) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:1.23E-02; Z-score:5.30E-01 | ||
|
Methylation in Case |
8.54E-01 (Median) | Methylation in Control | 8.34E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon62 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg25738529) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:1.40E-02; Z-score:-7.35E-01 | ||
|
Methylation in Case |
4.91E-01 (Median) | Methylation in Control | 5.29E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon63 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg19931596) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:3.04E-02; Z-score:1.72E-01 | ||
|
Methylation in Case |
8.47E-01 (Median) | Methylation in Control | 8.42E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon64 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg13270055) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:3.19E-02; Z-score:1.30E-01 | ||
|
Methylation in Case |
6.43E-01 (Median) | Methylation in Control | 6.33E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon65 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg12569736) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:3.43E-02; Z-score:4.51E-01 | ||
|
Methylation in Case |
3.57E-01 (Median) | Methylation in Control | 3.45E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon66 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg21853806) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:3.56E-02; Z-score:5.85E-01 | ||
|
Methylation in Case |
8.35E-01 (Median) | Methylation in Control | 8.25E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon67 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg10351795) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:3.58E-02; Z-score:6.53E-01 | ||
|
Methylation in Case |
6.99E-01 (Median) | Methylation in Control | 6.70E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon68 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
Body (cg03198012) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:4.56E-02; Z-score:1.22E+00 | ||
|
Methylation in Case |
7.84E-01 (Median) | Methylation in Control | 7.33E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon69 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
3'UTR (cg17442852) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:1.11E-06; Z-score:-1.25E+00 | ||
|
Methylation in Case |
7.53E-01 (Median) | Methylation in Control | 8.10E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon70 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
3'UTR (cg14729961) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:5.12E-04; Z-score:1.02E+00 | ||
|
Methylation in Case |
8.35E-01 (Median) | Methylation in Control | 8.16E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon71 |
Methylation of SLC12A7 in pancretic ductal adenocarcinoma | [ 2 ] | |||
|
Location |
3'UTR (cg03988700) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:3.69E-02; Z-score:-6.67E-01 | ||
|
Methylation in Case |
5.53E-01 (Median) | Methylation in Control | 5.99E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
21 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
5'UTR (cg07510333) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:9.18E-03; Z-score:2.41E+00 | ||
|
Methylation in Case |
9.38E-01 (Median) | Methylation in Control | 9.01E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
5'UTR (cg08310866) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.11E+00 | Statistic Test | p-value:3.19E-02; Z-score:7.19E+00 | ||
|
Methylation in Case |
6.16E-01 (Median) | Methylation in Control | 2.93E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg21455983) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:1.43E-03; Z-score:5.27E+00 | ||
|
Methylation in Case |
9.40E-01 (Median) | Methylation in Control | 9.00E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg09452751) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.48E+00 | Statistic Test | p-value:4.82E-03; Z-score:2.73E+00 | ||
|
Methylation in Case |
1.00E-01 (Median) | Methylation in Control | 6.74E-02 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg07992308) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:6.96E-03; Z-score:-4.20E+00 | ||
|
Methylation in Case |
4.00E-01 (Median) | Methylation in Control | 5.43E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg14442421) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:3.53E+00 | Statistic Test | p-value:7.75E-03; Z-score:9.77E+00 | ||
|
Methylation in Case |
4.34E-01 (Median) | Methylation in Control | 1.23E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg18939241) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:1.09E-02; Z-score:2.38E+00 | ||
|
Methylation in Case |
9.06E-01 (Median) | Methylation in Control | 7.91E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg03453890) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:3.80E-02; Z-score:1.84E+00 | ||
|
Methylation in Case |
8.58E-01 (Median) | Methylation in Control | 7.86E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
TSS200 (cg13407335) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.46E+00 | Statistic Test | p-value:1.53E-03; Z-score:5.36E+00 | ||
|
Methylation in Case |
7.78E-01 (Median) | Methylation in Control | 5.31E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
TSS200 (cg04574090) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:3.95E+00 | Statistic Test | p-value:1.60E-03; Z-score:1.88E+01 | ||
|
Methylation in Case |
7.09E-01 (Median) | Methylation in Control | 1.79E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
1stExon (cg07830847) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:5.23E-03; Z-score:3.29E+00 | ||
|
Methylation in Case |
9.53E-01 (Median) | Methylation in Control | 9.43E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
Body (cg14009098) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.66E+00 | Statistic Test | p-value:1.56E-03; Z-score:9.31E+00 | ||
|
Methylation in Case |
5.85E-01 (Median) | Methylation in Control | 2.20E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
Body (cg07567497) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:2.86E-03; Z-score:4.01E+00 | ||
|
Methylation in Case |
9.35E-01 (Median) | Methylation in Control | 9.07E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
Body (cg21690645) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:3.79E-03; Z-score:3.26E+00 | ||
|
Methylation in Case |
9.53E-01 (Median) | Methylation in Control | 8.72E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
Body (cg08362628) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.64E+00 | Statistic Test | p-value:7.09E-03; Z-score:2.79E+00 | ||
|
Methylation in Case |
7.38E-01 (Median) | Methylation in Control | 4.50E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
Body (cg25600103) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:1.58E-02; Z-score:2.76E+00 | ||
|
Methylation in Case |
7.08E-01 (Median) | Methylation in Control | 5.78E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
Body (cg11231434) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:1.92E-02; Z-score:2.50E+00 | ||
|
Methylation in Case |
9.28E-01 (Median) | Methylation in Control | 8.66E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
Body (cg03879320) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:3.26E-02; Z-score:1.76E+00 | ||
|
Methylation in Case |
9.01E-01 (Median) | Methylation in Control | 8.36E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
Body (cg04553355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:3.97E-02; Z-score:1.59E+00 | ||
|
Methylation in Case |
9.54E-01 (Median) | Methylation in Control | 9.31E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
Body (cg25925473) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:3.97E-02; Z-score:1.67E+00 | ||
|
Methylation in Case |
9.20E-01 (Median) | Methylation in Control | 8.71E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC12A7 in prostate cancer | [ 3 ] | |||
|
Location |
Body (cg02350636) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:4.27E-02; Z-score:2.36E+00 | ||
|
Methylation in Case |
9.52E-01 (Median) | Methylation in Control | 9.27E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
67 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg24000908) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.52E+00 | Statistic Test | p-value:1.06E-08; Z-score:-8.07E+00 | ||
|
Methylation in Case |
2.49E-01 (Median) | Methylation in Control | 6.28E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg02739870) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.65E+00 | Statistic Test | p-value:1.68E-08; Z-score:-8.07E+00 | ||
|
Methylation in Case |
2.35E-01 (Median) | Methylation in Control | 6.22E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
TSS200 (cg11962947) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:1.95E-02; Z-score:1.47E+00 | ||
|
Methylation in Case |
7.87E-02 (Median) | Methylation in Control | 6.73E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg18677871) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.48E+00 | Statistic Test | p-value:3.37E-13; Z-score:1.35E+01 | ||
|
Methylation in Case |
7.47E-01 (Median) | Methylation in Control | 5.06E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg01266985) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.38E+00 | Statistic Test | p-value:1.90E-10; Z-score:1.17E+01 | ||
|
Methylation in Case |
7.43E-01 (Median) | Methylation in Control | 5.38E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg23221540) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:2.99E-09; Z-score:8.22E+00 | ||
|
Methylation in Case |
8.53E-01 (Median) | Methylation in Control | 7.65E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg22172143) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.61E+00 | Statistic Test | p-value:5.22E-09; Z-score:-9.67E+00 | ||
|
Methylation in Case |
3.91E-01 (Median) | Methylation in Control | 6.31E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg12146829) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.78E+00 | Statistic Test | p-value:8.35E-09; Z-score:-6.49E+00 | ||
|
Methylation in Case |
3.38E-01 (Median) | Methylation in Control | 6.03E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg07459121) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:9.99E-08; Z-score:6.93E+00 | ||
|
Methylation in Case |
8.25E-01 (Median) | Methylation in Control | 6.85E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg00601711) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.39E+00 | Statistic Test | p-value:2.02E-06; Z-score:5.37E+00 | ||
|
Methylation in Case |
7.15E-01 (Median) | Methylation in Control | 5.17E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg18997983) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:2.07E-06; Z-score:5.65E+00 | ||
|
Methylation in Case |
8.08E-01 (Median) | Methylation in Control | 7.05E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg11677852) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:2.58E-06; Z-score:5.67E+00 | ||
|
Methylation in Case |
8.51E-01 (Median) | Methylation in Control | 7.90E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg08543327) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.23E+00 | Statistic Test | p-value:2.65E-06; Z-score:5.00E+00 | ||
|
Methylation in Case |
7.78E-01 (Median) | Methylation in Control | 6.32E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg00420510) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.29E+00 | Statistic Test | p-value:4.58E-06; Z-score:5.47E+00 | ||
|
Methylation in Case |
7.66E-01 (Median) | Methylation in Control | 5.93E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg10857489) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:4.99E-06; Z-score:-7.72E+00 | ||
|
Methylation in Case |
8.07E-01 (Median) | Methylation in Control | 8.93E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg06637017) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:6.04E-06; Z-score:5.48E+00 | ||
|
Methylation in Case |
8.25E-01 (Median) | Methylation in Control | 7.25E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg00346376) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:2.76E-05; Z-score:-4.52E+00 | ||
|
Methylation in Case |
7.03E-01 (Median) | Methylation in Control | 7.91E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg21946374) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:4.04E-05; Z-score:5.50E+00 | ||
|
Methylation in Case |
7.51E-01 (Median) | Methylation in Control | 6.72E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg15647725) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:4.38E-05; Z-score:9.05E+00 | ||
|
Methylation in Case |
5.86E-01 (Median) | Methylation in Control | 4.79E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg21334510) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.47E+00 | Statistic Test | p-value:9.07E-05; Z-score:-3.34E+00 | ||
|
Methylation in Case |
3.33E-01 (Median) | Methylation in Control | 4.91E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg23503101) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:1.08E-04; Z-score:-5.10E+00 | ||
|
Methylation in Case |
6.96E-01 (Median) | Methylation in Control | 7.86E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg21123417) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.87E+00 | Statistic Test | p-value:1.55E-04; Z-score:4.72E+00 | ||
|
Methylation in Case |
7.32E-01 (Median) | Methylation in Control | 3.91E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg03281154) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:3.02E-04; Z-score:-3.66E+00 | ||
|
Methylation in Case |
6.82E-01 (Median) | Methylation in Control | 7.61E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg13681701) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.67E+00 | Statistic Test | p-value:3.05E-04; Z-score:4.42E+00 | ||
|
Methylation in Case |
3.71E-01 (Median) | Methylation in Control | 2.23E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg02557110) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:5.41E-04; Z-score:4.20E+00 | ||
|
Methylation in Case |
8.58E-01 (Median) | Methylation in Control | 7.06E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg21516614) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:6.85E-04; Z-score:-5.37E+00 | ||
|
Methylation in Case |
9.28E-01 (Median) | Methylation in Control | 9.66E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg13301368) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:9.50E-04; Z-score:3.52E+00 | ||
|
Methylation in Case |
8.92E-01 (Median) | Methylation in Control | 8.24E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg01551729) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.20E-03; Z-score:-1.50E+00 | ||
|
Methylation in Case |
8.97E-01 (Median) | Methylation in Control | 9.08E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg00509649) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.68E+00 | Statistic Test | p-value:1.72E-03; Z-score:5.82E+00 | ||
|
Methylation in Case |
2.72E-01 (Median) | Methylation in Control | 1.62E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg02349468) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.54E+00 | Statistic Test | p-value:1.99E-03; Z-score:-2.11E+00 | ||
|
Methylation in Case |
4.28E-01 (Median) | Methylation in Control | 6.60E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg11577329) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.30E+00 | Statistic Test | p-value:2.60E-03; Z-score:-2.17E+00 | ||
|
Methylation in Case |
5.86E-01 (Median) | Methylation in Control | 7.59E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg05819249) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:2.96E-03; Z-score:2.88E+00 | ||
|
Methylation in Case |
8.73E-01 (Median) | Methylation in Control | 8.34E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg27072813) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:3.05E-03; Z-score:3.47E+00 | ||
|
Methylation in Case |
8.34E-01 (Median) | Methylation in Control | 7.81E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg14817049) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:3.20E-03; Z-score:-2.90E+00 | ||
|
Methylation in Case |
8.40E-01 (Median) | Methylation in Control | 8.85E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg14708411) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:4.76E-03; Z-score:-1.49E+00 | ||
|
Methylation in Case |
8.99E-01 (Median) | Methylation in Control | 9.14E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg16016716) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:5.06E-03; Z-score:4.27E+00 | ||
|
Methylation in Case |
8.16E-01 (Median) | Methylation in Control | 7.45E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon37 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg13592947) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.25E+00 | Statistic Test | p-value:6.56E-03; Z-score:-1.97E+00 | ||
|
Methylation in Case |
2.57E-01 (Median) | Methylation in Control | 3.21E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon38 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg11235297) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:6.95E-03; Z-score:2.65E+00 | ||
|
Methylation in Case |
8.21E-01 (Median) | Methylation in Control | 7.51E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon39 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg16419756) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:8.35E-03; Z-score:2.41E+00 | ||
|
Methylation in Case |
7.85E-01 (Median) | Methylation in Control | 7.22E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon40 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg23110957) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:8.88E-03; Z-score:2.11E+00 | ||
|
Methylation in Case |
8.68E-01 (Median) | Methylation in Control | 8.24E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon41 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg17733824) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.07E-02; Z-score:-1.05E+00 | ||
|
Methylation in Case |
9.28E-01 (Median) | Methylation in Control | 9.40E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon42 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg04467119) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.42E-02; Z-score:-1.35E+00 | ||
|
Methylation in Case |
7.67E-01 (Median) | Methylation in Control | 7.95E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon43 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg08293408) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.80E-02; Z-score:-1.48E+00 | ||
|
Methylation in Case |
8.28E-01 (Median) | Methylation in Control | 8.43E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon44 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg24928433) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:1.94E-02; Z-score:2.07E+00 | ||
|
Methylation in Case |
9.81E-01 (Median) | Methylation in Control | 9.21E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon45 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg26244838) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:2.14E-02; Z-score:-1.62E+00 | ||
|
Methylation in Case |
7.12E-01 (Median) | Methylation in Control | 7.67E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon46 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg04114636) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.27E-02; Z-score:-6.41E-01 | ||
|
Methylation in Case |
8.34E-01 (Median) | Methylation in Control | 8.41E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon47 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg25945676) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:2.40E-02; Z-score:1.70E+00 | ||
|
Methylation in Case |
8.59E-01 (Median) | Methylation in Control | 8.06E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon48 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg22673380) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:2.45E-02; Z-score:1.67E+00 | ||
|
Methylation in Case |
9.85E-01 (Median) | Methylation in Control | 9.73E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon49 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg06407917) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:2.56E-02; Z-score:-1.02E+00 | ||
|
Methylation in Case |
9.71E-01 (Median) | Methylation in Control | 9.73E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon50 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg10594510) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:2.88E-02; Z-score:2.02E+00 | ||
|
Methylation in Case |
7.93E-01 (Median) | Methylation in Control | 7.21E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon51 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg14047153) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.93E-02; Z-score:-8.16E-01 | ||
|
Methylation in Case |
9.61E-01 (Median) | Methylation in Control | 9.70E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon52 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg26213438) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:3.07E-02; Z-score:-1.54E+00 | ||
|
Methylation in Case |
7.14E-01 (Median) | Methylation in Control | 7.54E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon53 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg02079348) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.14E-02; Z-score:-5.84E-01 | ||
|
Methylation in Case |
6.74E-01 (Median) | Methylation in Control | 6.81E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon54 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg13376768) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:3.14E-02; Z-score:1.83E+00 | ||
|
Methylation in Case |
9.48E-01 (Median) | Methylation in Control | 9.33E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon55 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg16321301) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.23E-02; Z-score:-9.25E-01 | ||
|
Methylation in Case |
9.21E-01 (Median) | Methylation in Control | 9.26E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon56 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg16044603) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:3.39E-02; Z-score:1.57E+00 | ||
|
Methylation in Case |
8.23E-01 (Median) | Methylation in Control | 7.87E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon57 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg00697639) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:3.42E-02; Z-score:-1.12E+00 | ||
|
Methylation in Case |
7.85E-01 (Median) | Methylation in Control | 8.13E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon58 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg23491790) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:3.52E-02; Z-score:1.13E+00 | ||
|
Methylation in Case |
9.56E-01 (Median) | Methylation in Control | 9.40E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon59 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg22772691) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.24E+00 | Statistic Test | p-value:3.68E-02; Z-score:2.89E+00 | ||
|
Methylation in Case |
5.79E-01 (Median) | Methylation in Control | 4.67E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon60 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg23989709) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:3.83E-02; Z-score:-1.56E+00 | ||
|
Methylation in Case |
8.46E-01 (Median) | Methylation in Control | 8.62E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon61 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg11713480) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:4.00E-02; Z-score:-9.67E-01 | ||
|
Methylation in Case |
8.71E-01 (Median) | Methylation in Control | 8.85E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon62 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg06973176) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:4.37E-02; Z-score:3.00E+00 | ||
|
Methylation in Case |
7.86E-01 (Median) | Methylation in Control | 7.42E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon63 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg16374080) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:4.59E-02; Z-score:1.59E+00 | ||
|
Methylation in Case |
7.84E-01 (Median) | Methylation in Control | 7.22E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon64 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg11628781) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:4.79E-02; Z-score:-1.50E+00 | ||
|
Methylation in Case |
8.54E-01 (Median) | Methylation in Control | 8.70E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon65 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
Body (cg18576686) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:4.86E-02; Z-score:2.01E+00 | ||
|
Methylation in Case |
8.02E-01 (Median) | Methylation in Control | 7.58E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon66 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
3'UTR (cg10876737) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:2.07E-04; Z-score:-3.22E+00 | ||
|
Methylation in Case |
9.00E-01 (Median) | Methylation in Control | 9.57E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon67 |
Methylation of SLC12A7 in bladder cancer | [ 4 ] | |||
|
Location |
3'UTR (cg19086001) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:4.21E-02; Z-score:-1.76E+00 | ||
|
Methylation in Case |
7.04E-01 (Median) | Methylation in Control | 7.62E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
79 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg24000908) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.54E+00 | Statistic Test | p-value:8.70E-23; Z-score:-4.09E+00 | ||
|
Methylation in Case |
4.41E-01 (Median) | Methylation in Control | 6.80E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg02739870) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.53E+00 | Statistic Test | p-value:2.65E-20; Z-score:-4.38E+00 | ||
|
Methylation in Case |
4.18E-01 (Median) | Methylation in Control | 6.38E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg15083845) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.89E-03; Z-score:-7.71E-01 | ||
|
Methylation in Case |
7.65E-01 (Median) | Methylation in Control | 8.08E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
TSS200 (cg21985251) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.36E+00 | Statistic Test | p-value:1.96E-02; Z-score:5.78E-01 | ||
|
Methylation in Case |
4.60E-02 (Median) | Methylation in Control | 3.39E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg16374080) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:9.01E-19; Z-score:2.35E+00 | ||
|
Methylation in Case |
7.95E-01 (Median) | Methylation in Control | 6.69E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg21123417) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.33E+00 | Statistic Test | p-value:2.30E-18; Z-score:2.80E+00 | ||
|
Methylation in Case |
6.24E-01 (Median) | Methylation in Control | 4.68E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg18677871) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:2.29E-16; Z-score:2.37E+00 | ||
|
Methylation in Case |
7.14E-01 (Median) | Methylation in Control | 5.98E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg25655333) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:2.84E-16; Z-score:2.21E+00 | ||
|
Methylation in Case |
7.03E-01 (Median) | Methylation in Control | 5.88E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg16997104) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.18E+00 | Statistic Test | p-value:6.94E-16; Z-score:1.92E+00 | ||
|
Methylation in Case |
8.81E-01 (Median) | Methylation in Control | 7.47E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg25945676) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:1.25E-14; Z-score:1.87E+00 | ||
|
Methylation in Case |
8.37E-01 (Median) | Methylation in Control | 6.97E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg19854293) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.18E+00 | Statistic Test | p-value:9.29E-14; Z-score:2.02E+00 | ||
|
Methylation in Case |
8.04E-01 (Median) | Methylation in Control | 6.79E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg01266985) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:4.02E-12; Z-score:1.93E+00 | ||
|
Methylation in Case |
7.39E-01 (Median) | Methylation in Control | 6.52E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg22955595) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:2.01E-11; Z-score:2.04E+00 | ||
|
Methylation in Case |
7.73E-01 (Median) | Methylation in Control | 6.42E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg23491790) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:1.86E-09; Z-score:1.48E+00 | ||
|
Methylation in Case |
8.81E-01 (Median) | Methylation in Control | 7.21E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg27665580) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:2.11E-09; Z-score:1.33E+00 | ||
|
Methylation in Case |
6.90E-01 (Median) | Methylation in Control | 6.34E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg05978154) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.25E+00 | Statistic Test | p-value:6.49E-09; Z-score:1.87E+00 | ||
|
Methylation in Case |
8.53E-01 (Median) | Methylation in Control | 6.85E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg21513991) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:8.93E-09; Z-score:-1.50E+00 | ||
|
Methylation in Case |
7.77E-01 (Median) | Methylation in Control | 8.27E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg26500588) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:9.23E-09; Z-score:-1.31E+00 | ||
|
Methylation in Case |
7.46E-01 (Median) | Methylation in Control | 7.89E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg06973176) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:1.79E-08; Z-score:1.24E+00 | ||
|
Methylation in Case |
7.84E-01 (Median) | Methylation in Control | 7.07E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg09790628) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:6.31E-08; Z-score:1.01E+00 | ||
|
Methylation in Case |
8.39E-01 (Median) | Methylation in Control | 7.90E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg00509649) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.53E+00 | Statistic Test | p-value:2.16E-07; Z-score:2.04E+00 | ||
|
Methylation in Case |
2.91E-01 (Median) | Methylation in Control | 1.90E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg13301368) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:6.14E-07; Z-score:1.11E+00 | ||
|
Methylation in Case |
8.32E-01 (Median) | Methylation in Control | 7.66E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg15647725) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:9.19E-07; Z-score:1.69E+00 | ||
|
Methylation in Case |
6.35E-01 (Median) | Methylation in Control | 5.82E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg26220110) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.67E-06; Z-score:-1.12E+00 | ||
|
Methylation in Case |
7.96E-01 (Median) | Methylation in Control | 8.20E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg16163535) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:2.02E-06; Z-score:-1.19E+00 | ||
|
Methylation in Case |
8.35E-01 (Median) | Methylation in Control | 8.82E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg17969789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:2.40E-06; Z-score:1.67E+00 | ||
|
Methylation in Case |
7.45E-01 (Median) | Methylation in Control | 6.91E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg08830157) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:3.29E-06; Z-score:-1.11E+00 | ||
|
Methylation in Case |
5.90E-01 (Median) | Methylation in Control | 6.53E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg09951201) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:3.62E-06; Z-score:1.10E+00 | ||
|
Methylation in Case |
7.87E-01 (Median) | Methylation in Control | 7.19E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg13681701) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.33E+00 | Statistic Test | p-value:3.85E-06; Z-score:1.55E+00 | ||
|
Methylation in Case |
3.26E-01 (Median) | Methylation in Control | 2.45E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg00697639) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:4.75E-06; Z-score:-1.16E+00 | ||
|
Methylation in Case |
8.01E-01 (Median) | Methylation in Control | 8.33E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg00063291) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.19E-05; Z-score:-9.69E-01 | ||
|
Methylation in Case |
8.62E-01 (Median) | Methylation in Control | 8.97E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg11577329) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:5.72E-05; Z-score:-9.62E-01 | ||
|
Methylation in Case |
8.28E-01 (Median) | Methylation in Control | 8.59E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg11235297) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:7.40E-05; Z-score:-1.10E+00 | ||
|
Methylation in Case |
7.19E-01 (Median) | Methylation in Control | 8.02E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg10594510) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:7.58E-05; Z-score:-8.92E-01 | ||
|
Methylation in Case |
9.36E-01 (Median) | Methylation in Control | 9.56E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg21334510) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:1.09E-04; Z-score:1.06E+00 | ||
|
Methylation in Case |
6.47E-01 (Median) | Methylation in Control | 5.86E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg01171355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.34E-04; Z-score:-7.68E-01 | ||
|
Methylation in Case |
9.31E-01 (Median) | Methylation in Control | 9.45E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon37 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg05163933) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.35E-04; Z-score:-8.69E-01 | ||
|
Methylation in Case |
6.23E-01 (Median) | Methylation in Control | 6.63E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon38 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg26244838) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:1.68E-04; Z-score:1.45E+00 | ||
|
Methylation in Case |
7.85E-01 (Median) | Methylation in Control | 7.09E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon39 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg06344195) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:2.17E-04; Z-score:-6.83E-01 | ||
|
Methylation in Case |
8.49E-01 (Median) | Methylation in Control | 8.66E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon40 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg00420510) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:3.71E-04; Z-score:-8.88E-01 | ||
|
Methylation in Case |
6.85E-01 (Median) | Methylation in Control | 7.36E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon41 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg02557110) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.17E+00 | Statistic Test | p-value:4.24E-04; Z-score:-1.61E+00 | ||
|
Methylation in Case |
7.36E-01 (Median) | Methylation in Control | 8.64E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon42 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg06058262) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:6.99E-04; Z-score:-8.76E-01 | ||
|
Methylation in Case |
8.59E-01 (Median) | Methylation in Control | 8.81E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon43 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg17851021) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:8.51E-04; Z-score:-6.54E-01 | ||
|
Methylation in Case |
8.41E-01 (Median) | Methylation in Control | 8.63E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon44 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg10620395) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:1.01E-03; Z-score:-8.89E-01 | ||
|
Methylation in Case |
7.89E-01 (Median) | Methylation in Control | 8.32E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon45 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg23989709) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.58E-03; Z-score:-7.07E-01 | ||
|
Methylation in Case |
8.37E-01 (Median) | Methylation in Control | 8.51E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon46 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg14047153) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.85E-03; Z-score:-7.77E-01 | ||
|
Methylation in Case |
9.47E-01 (Median) | Methylation in Control | 9.70E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon47 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg16017429) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.05E-03; Z-score:-8.04E-01 | ||
|
Methylation in Case |
9.65E-01 (Median) | Methylation in Control | 9.78E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon48 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg14596589) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.43E-03; Z-score:-8.92E-01 | ||
|
Methylation in Case |
8.18E-01 (Median) | Methylation in Control | 8.61E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon49 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg23221540) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:2.55E-03; Z-score:2.06E-01 | ||
|
Methylation in Case |
8.21E-01 (Median) | Methylation in Control | 8.09E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon50 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg08293408) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:2.58E-03; Z-score:-7.46E-01 | ||
|
Methylation in Case |
8.18E-01 (Median) | Methylation in Control | 8.39E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon51 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg23115083) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:3.07E-03; Z-score:-9.92E-01 | ||
|
Methylation in Case |
8.00E-01 (Median) | Methylation in Control | 8.49E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon52 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg21946374) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:3.35E-03; Z-score:-8.66E-01 | ||
|
Methylation in Case |
7.01E-01 (Median) | Methylation in Control | 7.56E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon53 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg04184427) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:3.47E-03; Z-score:-5.77E-01 | ||
|
Methylation in Case |
6.76E-01 (Median) | Methylation in Control | 6.97E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon54 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg23503101) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:3.87E-03; Z-score:4.20E-01 | ||
|
Methylation in Case |
7.76E-01 (Median) | Methylation in Control | 7.38E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon55 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg25183683) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:4.01E-03; Z-score:4.45E-01 | ||
|
Methylation in Case |
8.15E-01 (Median) | Methylation in Control | 7.93E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon56 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg11628781) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:4.15E-03; Z-score:-7.98E-01 | ||
|
Methylation in Case |
8.26E-01 (Median) | Methylation in Control | 8.56E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon57 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg17097710) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.27E-03; Z-score:-6.74E-01 | ||
|
Methylation in Case |
8.92E-01 (Median) | Methylation in Control | 9.04E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon58 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg00095276) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.47E-03; Z-score:-3.08E-01 | ||
|
Methylation in Case |
8.22E-01 (Median) | Methylation in Control | 8.33E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon59 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg15601915) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:4.98E-03; Z-score:-7.28E-01 | ||
|
Methylation in Case |
7.47E-01 (Median) | Methylation in Control | 7.67E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon60 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg03281154) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:8.49E-03; Z-score:-5.96E-01 | ||
|
Methylation in Case |
7.23E-01 (Median) | Methylation in Control | 7.45E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon61 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg06637017) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:8.56E-03; Z-score:4.46E-01 | ||
|
Methylation in Case |
7.85E-01 (Median) | Methylation in Control | 7.65E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon62 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg16193717) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:8.81E-03; Z-score:5.02E-01 | ||
|
Methylation in Case |
8.18E-01 (Median) | Methylation in Control | 7.64E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon63 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg22772691) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:9.46E-03; Z-score:7.15E-01 | ||
|
Methylation in Case |
6.17E-01 (Median) | Methylation in Control | 5.68E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon64 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg08543327) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:1.21E-02; Z-score:-6.74E-01 | ||
|
Methylation in Case |
6.49E-01 (Median) | Methylation in Control | 7.18E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon65 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg22955555) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:1.34E-02; Z-score:7.71E-01 | ||
|
Methylation in Case |
7.87E-01 (Median) | Methylation in Control | 7.47E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon66 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg25388971) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:1.49E-02; Z-score:2.09E-01 | ||
|
Methylation in Case |
9.01E-01 (Median) | Methylation in Control | 8.96E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon67 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg19773466) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.90E-02; Z-score:-7.09E-01 | ||
|
Methylation in Case |
6.30E-01 (Median) | Methylation in Control | 6.69E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon68 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg00600029) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.91E-02; Z-score:-4.60E-01 | ||
|
Methylation in Case |
8.84E-01 (Median) | Methylation in Control | 8.93E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon69 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg02027730) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.50E-02; Z-score:-4.21E-01 | ||
|
Methylation in Case |
8.73E-01 (Median) | Methylation in Control | 8.82E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon70 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg10601043) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.61E-02; Z-score:-3.90E-01 | ||
|
Methylation in Case |
8.70E-01 (Median) | Methylation in Control | 8.79E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon71 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg18848012) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:2.81E-02; Z-score:-6.34E-01 | ||
|
Methylation in Case |
8.84E-01 (Median) | Methylation in Control | 9.02E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon72 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg16419756) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:3.00E-02; Z-score:-7.64E-01 | ||
|
Methylation in Case |
7.62E-01 (Median) | Methylation in Control | 7.95E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon73 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg20730619) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.25E-02; Z-score:-3.28E-01 | ||
|
Methylation in Case |
8.18E-01 (Median) | Methylation in Control | 8.26E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon74 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg27072813) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:4.50E-02; Z-score:-3.58E-01 | ||
|
Methylation in Case |
7.41E-01 (Median) | Methylation in Control | 7.65E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon75 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg27585120) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.53E-02; Z-score:-4.34E-01 | ||
|
Methylation in Case |
8.43E-01 (Median) | Methylation in Control | 8.54E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon76 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg08351607) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.57E-02; Z-score:-3.05E-01 | ||
|
Methylation in Case |
8.03E-01 (Median) | Methylation in Control | 8.09E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon77 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
Body (cg02079348) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:4.87E-02; Z-score:7.10E-01 | ||
|
Methylation in Case |
6.91E-01 (Median) | Methylation in Control | 6.63E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon78 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
3'UTR (cg10876737) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:2.76E-03; Z-score:-1.16E+00 | ||
|
Methylation in Case |
9.42E-01 (Median) | Methylation in Control | 9.72E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon79 |
Methylation of SLC12A7 in breast cancer | [ 5 ] | |||
|
Location |
3'UTR (cg17568547) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:3.75E-02; Z-score:-7.76E-01 | ||
|
Methylation in Case |
8.51E-01 (Median) | Methylation in Control | 8.77E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
74 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg24000908) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.43E+00 | Statistic Test | p-value:3.77E-13; Z-score:-4.84E+00 | ||
|
Methylation in Case |
6.02E-01 (Median) | Methylation in Control | 8.61E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg02739870) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.47E+00 | Statistic Test | p-value:1.27E-12; Z-score:-4.09E+00 | ||
|
Methylation in Case |
5.49E-01 (Median) | Methylation in Control | 8.10E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg15083845) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.15E-02; Z-score:-8.85E-01 | ||
|
Methylation in Case |
9.54E-01 (Median) | Methylation in Control | 9.64E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS200 (cg23091824) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:1.04E-03; Z-score:-9.68E-01 | ||
|
Methylation in Case |
2.84E-02 (Median) | Methylation in Control | 3.29E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
TSS200 (cg21985251) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:2.14E-02; Z-score:-1.11E+00 | ||
|
Methylation in Case |
3.75E-02 (Median) | Methylation in Control | 4.22E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg25970471) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:7.08E-11; Z-score:1.64E+00 | ||
|
Methylation in Case |
8.56E-01 (Median) | Methylation in Control | 8.02E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg16997104) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:9.39E-11; Z-score:2.04E+00 | ||
|
Methylation in Case |
9.57E-01 (Median) | Methylation in Control | 8.69E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg10620395) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.10E-10; Z-score:-2.30E+00 | ||
|
Methylation in Case |
9.13E-01 (Median) | Methylation in Control | 9.45E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg21513991) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:1.09E-09; Z-score:-4.16E+00 | ||
|
Methylation in Case |
8.92E-01 (Median) | Methylation in Control | 9.35E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg02039689) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:3.48E-09; Z-score:2.89E+00 | ||
|
Methylation in Case |
9.67E-01 (Median) | Methylation in Control | 8.68E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg25528709) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:6.31E-09; Z-score:1.26E+00 | ||
|
Methylation in Case |
9.80E-01 (Median) | Methylation in Control | 9.44E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg04226648) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.16E+00 | Statistic Test | p-value:1.94E-08; Z-score:2.83E+00 | ||
|
Methylation in Case |
9.75E-01 (Median) | Methylation in Control | 8.43E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg02079348) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:1.25E-07; Z-score:-3.77E+00 | ||
|
Methylation in Case |
7.44E-01 (Median) | Methylation in Control | 8.27E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg26500588) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:2.04E-07; Z-score:-2.49E+00 | ||
|
Methylation in Case |
8.81E-01 (Median) | Methylation in Control | 9.13E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg12146829) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.84E+00 | Statistic Test | p-value:1.39E-06; Z-score:-2.97E+00 | ||
|
Methylation in Case |
2.41E-01 (Median) | Methylation in Control | 4.44E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg17969789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:1.77E-06; Z-score:2.51E+00 | ||
|
Methylation in Case |
8.79E-01 (Median) | Methylation in Control | 7.89E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg08702805) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.89E-06; Z-score:-1.19E+00 | ||
|
Methylation in Case |
8.91E-01 (Median) | Methylation in Control | 9.12E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg13299707) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:2.20E-06; Z-score:-1.18E+00 | ||
|
Methylation in Case |
9.19E-01 (Median) | Methylation in Control | 9.34E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg00697639) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:3.65E-06; Z-score:-4.85E+00 | ||
|
Methylation in Case |
8.89E-01 (Median) | Methylation in Control | 9.40E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg06344195) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.75E-06; Z-score:-1.28E+00 | ||
|
Methylation in Case |
9.43E-01 (Median) | Methylation in Control | 9.55E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg00063291) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:5.91E-06; Z-score:-2.24E+00 | ||
|
Methylation in Case |
9.54E-01 (Median) | Methylation in Control | 9.75E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg03281154) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:7.69E-06; Z-score:-3.07E+00 | ||
|
Methylation in Case |
7.58E-01 (Median) | Methylation in Control | 8.45E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg18196463) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:5.25E-05; Z-score:-7.27E-01 | ||
|
Methylation in Case |
9.80E-01 (Median) | Methylation in Control | 9.82E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg10601043) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:5.57E-05; Z-score:-1.28E+00 | ||
|
Methylation in Case |
9.67E-01 (Median) | Methylation in Control | 9.74E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg04213775) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:7.78E-05; Z-score:-1.74E+00 | ||
|
Methylation in Case |
9.11E-01 (Median) | Methylation in Control | 9.32E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg00601711) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.37E+00 | Statistic Test | p-value:8.47E-05; Z-score:4.13E+00 | ||
|
Methylation in Case |
8.19E-01 (Median) | Methylation in Control | 5.99E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg15597069) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:9.35E-05; Z-score:-8.60E-01 | ||
|
Methylation in Case |
9.69E-01 (Median) | Methylation in Control | 9.74E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg22955595) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:2.86E-04; Z-score:-1.12E+00 | ||
|
Methylation in Case |
8.81E-01 (Median) | Methylation in Control | 8.98E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg11805188) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:3.71E-04; Z-score:1.16E+00 | ||
|
Methylation in Case |
5.38E-01 (Median) | Methylation in Control | 4.85E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg01171355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:3.96E-04; Z-score:-6.10E-01 | ||
|
Methylation in Case |
9.69E-01 (Median) | Methylation in Control | 9.72E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg08293408) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.33E-04; Z-score:-1.15E+00 | ||
|
Methylation in Case |
9.39E-01 (Median) | Methylation in Control | 9.50E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg22955555) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:4.39E-04; Z-score:-1.58E+00 | ||
|
Methylation in Case |
8.98E-01 (Median) | Methylation in Control | 9.22E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg26244838) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:5.13E-04; Z-score:-2.32E+00 | ||
|
Methylation in Case |
6.82E-01 (Median) | Methylation in Control | 7.65E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg04380229) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:6.77E-04; Z-score:-9.79E-01 | ||
|
Methylation in Case |
9.50E-01 (Median) | Methylation in Control | 9.59E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg17788850) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:7.32E-04; Z-score:-1.17E+00 | ||
|
Methylation in Case |
9.61E-01 (Median) | Methylation in Control | 9.70E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg00346376) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:8.53E-04; Z-score:-2.26E+00 | ||
|
Methylation in Case |
6.46E-01 (Median) | Methylation in Control | 7.36E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon37 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg14596589) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:8.61E-04; Z-score:-2.73E+00 | ||
|
Methylation in Case |
6.90E-01 (Median) | Methylation in Control | 7.90E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon38 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg09547427) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.01E-03; Z-score:-1.01E+00 | ||
|
Methylation in Case |
8.92E-01 (Median) | Methylation in Control | 9.11E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon39 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg11628781) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.25E-03; Z-score:-6.59E-01 | ||
|
Methylation in Case |
9.70E-01 (Median) | Methylation in Control | 9.74E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon40 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg08344943) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.34E-03; Z-score:-7.15E-01 | ||
|
Methylation in Case |
9.61E-01 (Median) | Methylation in Control | 9.63E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon41 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg07459121) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:1.95E-03; Z-score:1.36E+00 | ||
|
Methylation in Case |
7.86E-01 (Median) | Methylation in Control | 7.42E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon42 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg08351607) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.08E-03; Z-score:-5.49E-01 | ||
|
Methylation in Case |
9.09E-01 (Median) | Methylation in Control | 9.17E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon43 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg11713480) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.18E-03; Z-score:-1.51E+00 | ||
|
Methylation in Case |
9.77E-01 (Median) | Methylation in Control | 9.82E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon44 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg20730619) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.23E-03; Z-score:-1.40E+00 | ||
|
Methylation in Case |
9.25E-01 (Median) | Methylation in Control | 9.36E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon45 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg22673380) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:2.40E-03; Z-score:6.27E-01 | ||
|
Methylation in Case |
9.79E-01 (Median) | Methylation in Control | 9.72E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon46 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg05819249) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:2.45E-03; Z-score:-8.57E-01 | ||
|
Methylation in Case |
8.72E-01 (Median) | Methylation in Control | 8.94E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon47 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg14671982) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:3.02E-03; Z-score:-1.28E+00 | ||
|
Methylation in Case |
7.41E-01 (Median) | Methylation in Control | 7.81E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon48 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg21516614) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:3.65E-03; Z-score:-9.10E-01 | ||
|
Methylation in Case |
9.79E-01 (Median) | Methylation in Control | 9.81E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon49 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg23989709) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.81E-03; Z-score:-6.86E-01 | ||
|
Methylation in Case |
9.36E-01 (Median) | Methylation in Control | 9.43E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon50 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg03343453) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:5.67E-03; Z-score:-4.35E-01 | ||
|
Methylation in Case |
9.73E-01 (Median) | Methylation in Control | 9.76E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon51 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg18997983) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:7.57E-03; Z-score:-1.56E+00 | ||
|
Methylation in Case |
6.60E-01 (Median) | Methylation in Control | 7.25E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon52 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg24117063) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:7.71E-03; Z-score:1.66E+00 | ||
|
Methylation in Case |
9.02E-01 (Median) | Methylation in Control | 8.60E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon53 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg08830157) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.24E+00 | Statistic Test | p-value:7.75E-03; Z-score:-2.63E+00 | ||
|
Methylation in Case |
4.33E-01 (Median) | Methylation in Control | 5.35E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon54 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg21946374) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:8.60E-03; Z-score:-1.02E+00 | ||
|
Methylation in Case |
6.99E-01 (Median) | Methylation in Control | 7.37E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon55 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg02027730) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:9.83E-03; Z-score:-9.24E-01 | ||
|
Methylation in Case |
9.71E-01 (Median) | Methylation in Control | 9.77E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon56 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg19764541) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.11E-02; Z-score:-3.71E-01 | ||
|
Methylation in Case |
9.80E-01 (Median) | Methylation in Control | 9.81E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon57 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg23514135) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:1.22E-02; Z-score:-1.31E+00 | ||
|
Methylation in Case |
7.19E-01 (Median) | Methylation in Control | 7.85E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon58 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg23948452) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:1.29E-02; Z-score:8.44E-01 | ||
|
Methylation in Case |
9.52E-01 (Median) | Methylation in Control | 9.45E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon59 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg17097710) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.33E-02; Z-score:-1.82E-01 | ||
|
Methylation in Case |
9.75E-01 (Median) | Methylation in Control | 9.76E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon60 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg04467119) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.63E-02; Z-score:-8.27E-02 | ||
|
Methylation in Case |
8.92E-01 (Median) | Methylation in Control | 8.93E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon61 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg16017429) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.87E-02; Z-score:-3.01E-01 | ||
|
Methylation in Case |
9.81E-01 (Median) | Methylation in Control | 9.82E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon62 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg26439015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:1.87E-02; Z-score:2.61E-01 | ||
|
Methylation in Case |
1.23E-02 (Median) | Methylation in Control | 1.20E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon63 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg15647725) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:1.98E-02; Z-score:9.82E-01 | ||
|
Methylation in Case |
6.25E-01 (Median) | Methylation in Control | 5.94E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon64 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg02557110) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:2.01E-02; Z-score:4.72E-01 | ||
|
Methylation in Case |
9.22E-01 (Median) | Methylation in Control | 9.07E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon65 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg23998435) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:2.35E-02; Z-score:-2.01E-02 | ||
|
Methylation in Case |
9.69E-01 (Median) | Methylation in Control | 9.69E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon66 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg00600029) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:2.57E-02; Z-score:-3.68E-01 | ||
|
Methylation in Case |
9.62E-01 (Median) | Methylation in Control | 9.65E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon67 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg04114636) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.89E-02; Z-score:-4.34E-01 | ||
|
Methylation in Case |
9.39E-01 (Median) | Methylation in Control | 9.45E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon68 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg11677852) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:3.12E-02; Z-score:-1.73E+00 | ||
|
Methylation in Case |
6.90E-01 (Median) | Methylation in Control | 7.59E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon69 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg12199905) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:4.60E-02; Z-score:-3.23E-01 | ||
|
Methylation in Case |
9.73E-01 (Median) | Methylation in Control | 9.75E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon70 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg04184427) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.00E+00 | Statistic Test | p-value:4.83E-02; Z-score:4.49E-02 | ||
|
Methylation in Case |
8.69E-01 (Median) | Methylation in Control | 8.69E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon71 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg14981610) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:4.88E-02; Z-score:-4.18E-01 | ||
|
Methylation in Case |
9.50E-01 (Median) | Methylation in Control | 9.54E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon72 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
Body (cg10489284) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:4.92E-02; Z-score:1.35E+00 | ||
|
Methylation in Case |
9.66E-01 (Median) | Methylation in Control | 9.02E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon73 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
3'UTR (cg17568547) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.16E-07; Z-score:-1.55E+00 | ||
|
Methylation in Case |
9.65E-01 (Median) | Methylation in Control | 9.73E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon74 |
Methylation of SLC12A7 in clear cell renal cell carcinoma | [ 6 ] | |||
|
Location |
3'UTR (cg10876737) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.50E-04; Z-score:-1.75E+00 | ||
|
Methylation in Case |
9.68E-01 (Median) | Methylation in Control | 9.74E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
76 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg15083845) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:7.24E-09; Z-score:-2.33E+00 | ||
|
Methylation in Case |
8.61E-01 (Median) | Methylation in Control | 9.05E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg24000908) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:3.25E-07; Z-score:-1.55E+00 | ||
|
Methylation in Case |
4.73E-01 (Median) | Methylation in Control | 6.39E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg02739870) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.28E+00 | Statistic Test | p-value:4.73E-06; Z-score:-1.26E+00 | ||
|
Methylation in Case |
4.60E-01 (Median) | Methylation in Control | 5.88E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS200 (cg11962947) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.26E+00 | Statistic Test | p-value:2.77E-06; Z-score:1.52E+00 | ||
|
Methylation in Case |
4.79E-02 (Median) | Methylation in Control | 3.79E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
TSS200 (cg19524810) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.39E+00 | Statistic Test | p-value:8.29E-04; Z-score:-9.04E-01 | ||
|
Methylation in Case |
2.71E-02 (Median) | Methylation in Control | 3.78E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg19773466) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.25E+00 | Statistic Test | p-value:6.49E-10; Z-score:-2.08E+00 | ||
|
Methylation in Case |
5.17E-01 (Median) | Methylation in Control | 6.45E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg21123417) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.28E+00 | Statistic Test | p-value:1.16E-09; Z-score:2.22E+00 | ||
|
Methylation in Case |
6.75E-01 (Median) | Methylation in Control | 5.27E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg16163535) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.19E+00 | Statistic Test | p-value:2.10E-08; Z-score:-2.29E+00 | ||
|
Methylation in Case |
7.35E-01 (Median) | Methylation in Control | 8.72E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg14596589) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:1.07E-07; Z-score:-1.89E+00 | ||
|
Methylation in Case |
7.71E-01 (Median) | Methylation in Control | 8.83E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg00420510) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.18E+00 | Statistic Test | p-value:8.76E-07; Z-score:1.88E+00 | ||
|
Methylation in Case |
8.54E-01 (Median) | Methylation in Control | 7.26E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg08830157) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.28E+00 | Statistic Test | p-value:9.58E-07; Z-score:-1.66E+00 | ||
|
Methylation in Case |
4.80E-01 (Median) | Methylation in Control | 6.14E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg10857489) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:1.48E-06; Z-score:-1.43E+00 | ||
|
Methylation in Case |
8.62E-01 (Median) | Methylation in Control | 9.02E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg11235297) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:2.51E-06; Z-score:-1.56E+00 | ||
|
Methylation in Case |
8.03E-01 (Median) | Methylation in Control | 8.71E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg21946374) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:5.44E-06; Z-score:-1.06E+00 | ||
|
Methylation in Case |
8.28E-01 (Median) | Methylation in Control | 8.64E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg01171355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:5.91E-06; Z-score:-1.58E+00 | ||
|
Methylation in Case |
9.58E-01 (Median) | Methylation in Control | 9.69E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg00215425) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:1.49E-05; Z-score:9.62E-01 | ||
|
Methylation in Case |
8.51E-01 (Median) | Methylation in Control | 7.76E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg08516247) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.31E-05; Z-score:-1.45E+00 | ||
|
Methylation in Case |
9.53E-01 (Median) | Methylation in Control | 9.63E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg23110957) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:2.61E-05; Z-score:1.24E+00 | ||
|
Methylation in Case |
9.28E-01 (Median) | Methylation in Control | 9.04E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg18677871) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:3.35E-05; Z-score:1.36E+00 | ||
|
Methylation in Case |
8.33E-01 (Median) | Methylation in Control | 7.70E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg02557110) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:4.07E-05; Z-score:1.32E+00 | ||
|
Methylation in Case |
9.19E-01 (Median) | Methylation in Control | 8.49E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg00601711) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:6.63E-05; Z-score:1.38E+00 | ||
|
Methylation in Case |
8.02E-01 (Median) | Methylation in Control | 7.01E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg04467119) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:7.11E-05; Z-score:-1.05E+00 | ||
|
Methylation in Case |
8.85E-01 (Median) | Methylation in Control | 9.00E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg08543327) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:7.92E-05; Z-score:-8.00E-01 | ||
|
Methylation in Case |
5.97E-01 (Median) | Methylation in Control | 6.71E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg23503101) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:8.28E-05; Z-score:-1.02E+00 | ||
|
Methylation in Case |
8.74E-01 (Median) | Methylation in Control | 8.95E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg06637017) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:1.16E-04; Z-score:1.26E+00 | ||
|
Methylation in Case |
9.21E-01 (Median) | Methylation in Control | 8.98E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg10594510) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.53E+00 | Statistic Test | p-value:1.18E-04; Z-score:1.51E+00 | ||
|
Methylation in Case |
7.18E-01 (Median) | Methylation in Control | 4.69E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg00697639) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.25E-04; Z-score:-7.62E-01 | ||
|
Methylation in Case |
8.99E-01 (Median) | Methylation in Control | 9.09E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg18196463) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.57E-04; Z-score:-1.64E+00 | ||
|
Methylation in Case |
9.72E-01 (Median) | Methylation in Control | 9.77E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg11805188) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.25E+00 | Statistic Test | p-value:1.82E-04; Z-score:1.35E+00 | ||
|
Methylation in Case |
6.46E-01 (Median) | Methylation in Control | 5.16E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg13681701) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:1.88E-04; Z-score:7.45E-01 | ||
|
Methylation in Case |
3.54E-01 (Median) | Methylation in Control | 2.98E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg16321301) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.12E-04; Z-score:-1.45E+00 | ||
|
Methylation in Case |
9.50E-01 (Median) | Methylation in Control | 9.57E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg23989709) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.15E-04; Z-score:-1.14E+00 | ||
|
Methylation in Case |
9.24E-01 (Median) | Methylation in Control | 9.32E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg06344195) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:5.06E-04; Z-score:-1.07E+00 | ||
|
Methylation in Case |
9.37E-01 (Median) | Methylation in Control | 9.44E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg21516614) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:5.21E-04; Z-score:-1.08E+00 | ||
|
Methylation in Case |
9.55E-01 (Median) | Methylation in Control | 9.63E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg08293408) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:7.12E-04; Z-score:-5.84E-01 | ||
|
Methylation in Case |
9.36E-01 (Median) | Methylation in Control | 9.40E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg10489284) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:8.22E-04; Z-score:1.10E+00 | ||
|
Methylation in Case |
9.57E-01 (Median) | Methylation in Control | 9.46E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon37 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg23221540) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:1.05E-03; Z-score:1.19E+00 | ||
|
Methylation in Case |
9.39E-01 (Median) | Methylation in Control | 9.23E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon38 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg16044603) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.46E-03; Z-score:-6.14E-01 | ||
|
Methylation in Case |
9.14E-01 (Median) | Methylation in Control | 9.21E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon39 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg09050160) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:1.47E-03; Z-score:-1.03E+00 | ||
|
Methylation in Case |
5.84E-01 (Median) | Methylation in Control | 6.75E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon40 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg07459121) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:1.88E-03; Z-score:6.07E-01 | ||
|
Methylation in Case |
8.41E-01 (Median) | Methylation in Control | 8.15E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon41 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg22772691) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.27E+00 | Statistic Test | p-value:2.18E-03; Z-score:5.61E-01 | ||
|
Methylation in Case |
3.15E-01 (Median) | Methylation in Control | 2.48E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon42 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg23491790) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:2.54E-03; Z-score:1.02E+00 | ||
|
Methylation in Case |
9.09E-01 (Median) | Methylation in Control | 8.69E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon43 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg20730619) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.00E+00 | Statistic Test | p-value:2.76E-03; Z-score:3.54E-02 | ||
|
Methylation in Case |
9.33E-01 (Median) | Methylation in Control | 9.33E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon44 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg03281154) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:3.26E-03; Z-score:-7.38E-01 | ||
|
Methylation in Case |
8.54E-01 (Median) | Methylation in Control | 8.72E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon45 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg13299707) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.30E-03; Z-score:-6.98E-01 | ||
|
Methylation in Case |
9.19E-01 (Median) | Methylation in Control | 9.30E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon46 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg23998435) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.50E-03; Z-score:-1.65E+00 | ||
|
Methylation in Case |
9.54E-01 (Median) | Methylation in Control | 9.63E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon47 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg15601915) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.79E-03; Z-score:-5.03E-01 | ||
|
Methylation in Case |
8.96E-01 (Median) | Methylation in Control | 9.02E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon48 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg00098175) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:3.88E-03; Z-score:6.07E-02 | ||
|
Methylation in Case |
8.69E-01 (Median) | Methylation in Control | 8.45E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon49 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg11628781) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.90E-03; Z-score:-7.30E-01 | ||
|
Methylation in Case |
9.31E-01 (Median) | Methylation in Control | 9.40E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon50 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg24928433) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:4.86E-03; Z-score:1.05E+00 | ||
|
Methylation in Case |
9.83E-01 (Median) | Methylation in Control | 9.76E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon51 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg25655333) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:5.75E-03; Z-score:-5.59E-01 | ||
|
Methylation in Case |
8.15E-01 (Median) | Methylation in Control | 8.31E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon52 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg26213438) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:5.81E-03; Z-score:-6.02E-01 | ||
|
Methylation in Case |
8.41E-01 (Median) | Methylation in Control | 8.58E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon53 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg27665580) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:8.61E-03; Z-score:-8.02E-01 | ||
|
Methylation in Case |
8.37E-01 (Median) | Methylation in Control | 8.60E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon54 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg15647725) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:8.88E-03; Z-score:1.25E+00 | ||
|
Methylation in Case |
7.72E-01 (Median) | Methylation in Control | 7.35E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon55 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg11713480) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:8.91E-03; Z-score:-7.84E-01 | ||
|
Methylation in Case |
9.35E-01 (Median) | Methylation in Control | 9.42E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon56 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg17788850) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.00E+00 | Statistic Test | p-value:1.04E-02; Z-score:1.97E-01 | ||
|
Methylation in Case |
9.57E-01 (Median) | Methylation in Control | 9.55E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon57 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg08351607) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.16E-02; Z-score:-8.79E-02 | ||
|
Methylation in Case |
9.22E-01 (Median) | Methylation in Control | 9.23E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon58 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg19764541) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.37E-02; Z-score:-8.15E-02 | ||
|
Methylation in Case |
9.53E-01 (Median) | Methylation in Control | 9.53E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon59 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg17969789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.70E-02; Z-score:-5.24E-01 | ||
|
Methylation in Case |
8.84E-01 (Median) | Methylation in Control | 8.92E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon60 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg10620395) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.74E-02; Z-score:-7.67E-02 | ||
|
Methylation in Case |
9.15E-01 (Median) | Methylation in Control | 9.16E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon61 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg14817049) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.92E-02; Z-score:-7.97E-01 | ||
|
Methylation in Case |
9.13E-01 (Median) | Methylation in Control | 9.22E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon62 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg21513991) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.00E-02; Z-score:-3.82E-01 | ||
|
Methylation in Case |
9.09E-01 (Median) | Methylation in Control | 9.14E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon63 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg16246240) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:2.00E-02; Z-score:-3.88E-01 | ||
|
Methylation in Case |
7.46E-01 (Median) | Methylation in Control | 7.61E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon64 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg16997104) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:2.54E-02; Z-score:1.06E+00 | ||
|
Methylation in Case |
9.04E-01 (Median) | Methylation in Control | 8.65E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon65 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg00509649) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:2.56E-02; Z-score:3.50E-01 | ||
|
Methylation in Case |
2.88E-01 (Median) | Methylation in Control | 2.69E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon66 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg23514135) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:2.83E-02; Z-score:8.80E-01 | ||
|
Methylation in Case |
7.95E-01 (Median) | Methylation in Control | 6.99E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon67 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg18997983) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:3.09E-02; Z-score:2.29E-01 | ||
|
Methylation in Case |
8.64E-01 (Median) | Methylation in Control | 8.54E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon68 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg08702805) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:3.49E-02; Z-score:-1.06E-01 | ||
|
Methylation in Case |
9.10E-01 (Median) | Methylation in Control | 9.11E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon69 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg14671982) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.71E-02; Z-score:-1.98E-01 | ||
|
Methylation in Case |
8.51E-01 (Median) | Methylation in Control | 8.56E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon70 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg05163933) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:3.82E-02; Z-score:7.44E-01 | ||
|
Methylation in Case |
6.32E-01 (Median) | Methylation in Control | 5.60E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon71 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg27010096) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:3.84E-02; Z-score:5.18E-01 | ||
|
Methylation in Case |
9.14E-01 (Median) | Methylation in Control | 8.91E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon72 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg10601043) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:4.04E-02; Z-score:-2.21E-01 | ||
|
Methylation in Case |
9.47E-01 (Median) | Methylation in Control | 9.48E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon73 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg04184427) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:4.10E-02; Z-score:-1.23E-02 | ||
|
Methylation in Case |
8.68E-01 (Median) | Methylation in Control | 8.68E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon74 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
Body (cg22955595) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:4.68E-02; Z-score:-1.70E-01 | ||
|
Methylation in Case |
9.02E-01 (Median) | Methylation in Control | 9.05E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon75 |
Methylation of SLC12A7 in colorectal cancer | [ 7 ] | |||
|
Location |
3'UTR (cg19086001) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.33E-02; Z-score:-5.42E-01 | ||
|
Methylation in Case |
8.84E-01 (Median) | Methylation in Control | 8.93E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
100 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS1500 (cg03561565) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.79E+00 | Statistic Test | p-value:2.68E-20; Z-score:4.63E+00 | ||
|
Methylation in Case |
6.72E-01 (Median) | Methylation in Control | 3.76E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS1500 (cg25711246) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.42E+00 | Statistic Test | p-value:1.09E-16; Z-score:-2.68E+00 | ||
|
Methylation in Case |
4.09E-01 (Median) | Methylation in Control | 5.82E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS1500 (cg04589881) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.30E+00 | Statistic Test | p-value:9.00E-12; Z-score:-1.14E+01 | ||
|
Methylation in Case |
6.55E-01 (Median) | Methylation in Control | 8.50E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS1500 (cg07748193) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:5.65E-11; Z-score:-1.61E+00 | ||
|
Methylation in Case |
5.90E-01 (Median) | Methylation in Control | 6.76E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS1500 (cg12985923) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:3.19E-10; Z-score:-5.09E+00 | ||
|
Methylation in Case |
7.57E-01 (Median) | Methylation in Control | 8.79E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS1500 (cg26317549) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.46E+00 | Statistic Test | p-value:5.53E-10; Z-score:-3.72E+00 | ||
|
Methylation in Case |
4.19E-01 (Median) | Methylation in Control | 6.12E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS1500 (cg02739870) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:5.18E-05; Z-score:-1.12E+00 | ||
|
Methylation in Case |
5.53E-01 (Median) | Methylation in Control | 6.40E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS1500 (cg15083845) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.10E-04; Z-score:-8.85E-01 | ||
|
Methylation in Case |
7.92E-01 (Median) | Methylation in Control | 8.08E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS1500 (cg24000908) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:4.63E-03; Z-score:-4.55E-01 | ||
|
Methylation in Case |
6.00E-01 (Median) | Methylation in Control | 6.33E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg04431946) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.80E+00 | Statistic Test | p-value:3.06E-16; Z-score:6.77E+00 | ||
|
Methylation in Case |
4.52E-01 (Median) | Methylation in Control | 1.62E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg22988581) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.47E+00 | Statistic Test | p-value:9.85E-12; Z-score:-2.15E+00 | ||
|
Methylation in Case |
2.88E-01 (Median) | Methylation in Control | 4.23E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg25811526) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.26E+00 | Statistic Test | p-value:3.51E-11; Z-score:-1.98E+00 | ||
|
Methylation in Case |
4.34E-01 (Median) | Methylation in Control | 5.46E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg01325409) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.42E+00 | Statistic Test | p-value:3.40E-10; Z-score:-2.68E+00 | ||
|
Methylation in Case |
3.48E-01 (Median) | Methylation in Control | 4.96E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg21985251) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:1.40E-02; Z-score:-2.67E-01 | ||
|
Methylation in Case |
5.28E-02 (Median) | Methylation in Control | 6.11E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
TSS200 (cg23091824) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:2.77E-02; Z-score:-3.32E-01 | ||
|
Methylation in Case |
4.83E-02 (Median) | Methylation in Control | 5.61E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
1stExon (cg06946880) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.27E+00 | Statistic Test | p-value:2.43E-10; Z-score:-5.36E+00 | ||
|
Methylation in Case |
6.15E-01 (Median) | Methylation in Control | 7.80E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg14486338) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.53E+00 | Statistic Test | p-value:1.02E-28; Z-score:4.55E+00 | ||
|
Methylation in Case |
6.94E-01 (Median) | Methylation in Control | 2.75E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg02471153) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.82E+00 | Statistic Test | p-value:7.72E-24; Z-score:3.88E+00 | ||
|
Methylation in Case |
5.63E-01 (Median) | Methylation in Control | 3.10E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg24602704) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.61E+00 | Statistic Test | p-value:8.60E-23; Z-score:-1.26E+01 | ||
|
Methylation in Case |
3.29E-01 (Median) | Methylation in Control | 8.58E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg14718495) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.86E+00 | Statistic Test | p-value:1.68E-21; Z-score:-4.90E+00 | ||
|
Methylation in Case |
3.69E-01 (Median) | Methylation in Control | 6.86E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg21403761) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.74E+00 | Statistic Test | p-value:2.71E-21; Z-score:-6.26E+00 | ||
|
Methylation in Case |
3.59E-01 (Median) | Methylation in Control | 6.27E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg14790078) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.39E+00 | Statistic Test | p-value:1.08E-18; Z-score:-4.89E+00 | ||
|
Methylation in Case |
5.85E-01 (Median) | Methylation in Control | 8.13E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg13298691) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.69E+00 | Statistic Test | p-value:3.73E-18; Z-score:-4.89E+00 | ||
|
Methylation in Case |
5.17E-01 (Median) | Methylation in Control | 8.75E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg17526887) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.62E+00 | Statistic Test | p-value:1.77E-17; Z-score:-2.35E+00 | ||
|
Methylation in Case |
3.41E-01 (Median) | Methylation in Control | 5.54E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg20696050) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.34E+00 | Statistic Test | p-value:1.17E-15; Z-score:-1.56E+01 | ||
|
Methylation in Case |
6.41E-01 (Median) | Methylation in Control | 8.61E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg01949837) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.44E+00 | Statistic Test | p-value:4.62E-15; Z-score:-1.03E+01 | ||
|
Methylation in Case |
6.08E-01 (Median) | Methylation in Control | 8.75E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg05005358) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.65E+00 | Statistic Test | p-value:8.73E-15; Z-score:-1.11E+01 | ||
|
Methylation in Case |
4.83E-01 (Median) | Methylation in Control | 7.95E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg01316792) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:4.66E-14; Z-score:-8.99E+00 | ||
|
Methylation in Case |
5.77E-01 (Median) | Methylation in Control | 7.84E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg23617640) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.25E+00 | Statistic Test | p-value:5.37E-14; Z-score:-6.66E+00 | ||
|
Methylation in Case |
7.06E-01 (Median) | Methylation in Control | 8.82E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg22708635) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.37E+00 | Statistic Test | p-value:1.12E-13; Z-score:-7.23E+00 | ||
|
Methylation in Case |
6.30E-01 (Median) | Methylation in Control | 8.62E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg13890552) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.41E+00 | Statistic Test | p-value:1.22E-13; Z-score:-2.15E+00 | ||
|
Methylation in Case |
5.84E-01 (Median) | Methylation in Control | 8.23E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg08131081) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.33E+00 | Statistic Test | p-value:2.76E-13; Z-score:-1.12E+01 | ||
|
Methylation in Case |
6.96E-01 (Median) | Methylation in Control | 9.29E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg12845051) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:3.19E-13; Z-score:-9.43E+00 | ||
|
Methylation in Case |
5.88E-01 (Median) | Methylation in Control | 7.94E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg04602747) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.65E+00 | Statistic Test | p-value:5.14E-13; Z-score:-2.31E+00 | ||
|
Methylation in Case |
1.55E-01 (Median) | Methylation in Control | 2.55E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg14491776) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.33E+00 | Statistic Test | p-value:6.18E-12; Z-score:-3.72E+00 | ||
|
Methylation in Case |
6.53E-01 (Median) | Methylation in Control | 8.70E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg22334665) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.74E+00 | Statistic Test | p-value:1.27E-11; Z-score:2.11E+00 | ||
|
Methylation in Case |
4.09E-01 (Median) | Methylation in Control | 2.36E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon37 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg13603332) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.42E+00 | Statistic Test | p-value:3.23E-11; Z-score:-1.33E+00 | ||
|
Methylation in Case |
3.26E-01 (Median) | Methylation in Control | 4.64E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon38 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg24474319) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:3.62E-11; Z-score:-3.20E+00 | ||
|
Methylation in Case |
3.73E-01 (Median) | Methylation in Control | 5.07E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon39 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg24102222) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:3.77E-11; Z-score:1.39E+00 | ||
|
Methylation in Case |
7.45E-01 (Median) | Methylation in Control | 6.38E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon40 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg16619764) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.33E+00 | Statistic Test | p-value:4.94E-11; Z-score:-1.97E+00 | ||
|
Methylation in Case |
3.60E-01 (Median) | Methylation in Control | 4.78E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon41 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg18190534) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:6.60E-10; Z-score:-7.35E+00 | ||
|
Methylation in Case |
8.26E-01 (Median) | Methylation in Control | 9.59E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon42 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg13512859) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.21E+00 | Statistic Test | p-value:1.03E-09; Z-score:-1.76E+00 | ||
|
Methylation in Case |
4.45E-01 (Median) | Methylation in Control | 5.37E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon43 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg15383972) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:1.08E-09; Z-score:-4.89E+00 | ||
|
Methylation in Case |
8.46E-01 (Median) | Methylation in Control | 9.55E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon44 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg15647725) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:5.57E-09; Z-score:1.89E+00 | ||
|
Methylation in Case |
5.62E-01 (Median) | Methylation in Control | 5.00E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon45 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg26213438) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:6.03E-09; Z-score:1.59E+00 | ||
|
Methylation in Case |
7.09E-01 (Median) | Methylation in Control | 6.09E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon46 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg12146829) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.57E+00 | Statistic Test | p-value:6.29E-09; Z-score:-1.46E+00 | ||
|
Methylation in Case |
1.34E-01 (Median) | Methylation in Control | 2.10E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon47 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg05978154) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.29E+00 | Statistic Test | p-value:9.48E-09; Z-score:1.87E+00 | ||
|
Methylation in Case |
8.64E-01 (Median) | Methylation in Control | 6.71E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon48 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg22955595) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:1.63E-08; Z-score:9.55E-01 | ||
|
Methylation in Case |
7.61E-01 (Median) | Methylation in Control | 6.88E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon49 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg22172143) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.50E+00 | Statistic Test | p-value:1.94E-08; Z-score:-1.35E+00 | ||
|
Methylation in Case |
1.73E-01 (Median) | Methylation in Control | 2.59E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon50 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg08830157) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.23E+00 | Statistic Test | p-value:2.07E-07; Z-score:-1.17E+00 | ||
|
Methylation in Case |
2.43E-01 (Median) | Methylation in Control | 2.99E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon51 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg26270832) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:4.69E-07; Z-score:8.95E-01 | ||
|
Methylation in Case |
8.40E-01 (Median) | Methylation in Control | 7.41E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon52 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg27665580) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:6.13E-07; Z-score:-1.07E+00 | ||
|
Methylation in Case |
7.39E-01 (Median) | Methylation in Control | 7.72E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon53 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg14671982) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:1.68E-06; Z-score:1.46E+00 | ||
|
Methylation in Case |
6.89E-01 (Median) | Methylation in Control | 6.22E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon54 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg26500588) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:4.65E-06; Z-score:1.17E+00 | ||
|
Methylation in Case |
7.91E-01 (Median) | Methylation in Control | 7.48E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon55 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg25945676) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:6.02E-06; Z-score:1.12E+00 | ||
|
Methylation in Case |
8.77E-01 (Median) | Methylation in Control | 8.44E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon56 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg14596589) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.21E+00 | Statistic Test | p-value:1.14E-05; Z-score:-9.71E-01 | ||
|
Methylation in Case |
4.17E-01 (Median) | Methylation in Control | 5.05E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon57 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg02382320) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:2.27E-05; Z-score:1.17E+00 | ||
|
Methylation in Case |
9.04E-01 (Median) | Methylation in Control | 8.35E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon58 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg06973176) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:4.38E-05; Z-score:6.71E-01 | ||
|
Methylation in Case |
7.97E-01 (Median) | Methylation in Control | 7.82E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon59 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg09790628) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:4.70E-05; Z-score:1.20E+00 | ||
|
Methylation in Case |
8.20E-01 (Median) | Methylation in Control | 7.36E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon60 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg01551729) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.29E+00 | Statistic Test | p-value:5.60E-05; Z-score:-1.10E+00 | ||
|
Methylation in Case |
4.76E-01 (Median) | Methylation in Control | 6.11E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon61 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg13301368) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:5.75E-05; Z-score:9.21E-01 | ||
|
Methylation in Case |
8.90E-01 (Median) | Methylation in Control | 8.75E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon62 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg00063291) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:9.07E-05; Z-score:8.66E-01 | ||
|
Methylation in Case |
8.72E-01 (Median) | Methylation in Control | 8.45E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon63 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg25655333) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:9.15E-05; Z-score:-9.68E-01 | ||
|
Methylation in Case |
6.99E-01 (Median) | Methylation in Control | 7.25E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon64 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg13592947) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.31E+00 | Statistic Test | p-value:1.15E-04; Z-score:-1.27E+00 | ||
|
Methylation in Case |
1.97E-01 (Median) | Methylation in Control | 2.58E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon65 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg00095276) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:1.47E-04; Z-score:4.70E-01 | ||
|
Methylation in Case |
8.23E-01 (Median) | Methylation in Control | 8.09E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon66 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg16163535) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:2.26E-04; Z-score:-5.95E-01 | ||
|
Methylation in Case |
4.75E-01 (Median) | Methylation in Control | 5.52E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon67 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg03281154) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.78E-04; Z-score:-1.14E+00 | ||
|
Methylation in Case |
7.11E-01 (Median) | Methylation in Control | 7.46E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon68 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg04467119) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:3.38E-04; Z-score:3.48E-01 | ||
|
Methylation in Case |
7.67E-01 (Median) | Methylation in Control | 7.34E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon69 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg23115083) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:3.65E-04; Z-score:6.34E-01 | ||
|
Methylation in Case |
8.54E-01 (Median) | Methylation in Control | 8.33E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon70 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg10857489) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:4.89E-04; Z-score:-4.01E-01 | ||
|
Methylation in Case |
8.82E-01 (Median) | Methylation in Control | 8.98E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon71 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg16246240) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:8.16E-04; Z-score:6.58E-01 | ||
|
Methylation in Case |
6.50E-01 (Median) | Methylation in Control | 5.94E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon72 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg10594510) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.30E+00 | Statistic Test | p-value:8.62E-04; Z-score:1.59E+00 | ||
|
Methylation in Case |
6.85E-01 (Median) | Methylation in Control | 5.29E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon73 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg18576686) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:9.43E-04; Z-score:6.81E-01 | ||
|
Methylation in Case |
8.18E-01 (Median) | Methylation in Control | 7.97E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon74 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg26220110) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.20E-03; Z-score:-6.07E-01 | ||
|
Methylation in Case |
7.82E-01 (Median) | Methylation in Control | 7.94E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon75 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg00346376) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.17E+00 | Statistic Test | p-value:1.54E-03; Z-score:-1.04E+00 | ||
|
Methylation in Case |
4.70E-01 (Median) | Methylation in Control | 5.49E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon76 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg13681701) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.33E+00 | Statistic Test | p-value:1.55E-03; Z-score:-1.08E+00 | ||
|
Methylation in Case |
1.81E-01 (Median) | Methylation in Control | 2.40E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon77 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg09951201) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:1.62E-03; Z-score:9.78E-01 | ||
|
Methylation in Case |
7.31E-01 (Median) | Methylation in Control | 6.53E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon78 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg05636015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:3.21E-03; Z-score:5.42E-01 | ||
|
Methylation in Case |
8.27E-01 (Median) | Methylation in Control | 8.15E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon79 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg03343453) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:3.22E-03; Z-score:3.05E-01 | ||
|
Methylation in Case |
8.73E-01 (Median) | Methylation in Control | 8.69E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon80 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg08702805) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:3.89E-03; Z-score:6.75E-01 | ||
|
Methylation in Case |
8.02E-01 (Median) | Methylation in Control | 7.81E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon81 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg17733824) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.26E+00 | Statistic Test | p-value:4.53E-03; Z-score:-1.04E+00 | ||
|
Methylation in Case |
5.00E-01 (Median) | Methylation in Control | 6.28E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon82 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg26824947) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:5.88E-03; Z-score:3.17E-01 | ||
|
Methylation in Case |
6.93E-01 (Median) | Methylation in Control | 6.61E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon83 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg13299707) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:7.57E-03; Z-score:6.26E-01 | ||
|
Methylation in Case |
8.46E-01 (Median) | Methylation in Control | 8.33E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon84 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg02349468) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:1.03E-02; Z-score:3.84E-01 | ||
|
Methylation in Case |
5.63E-01 (Median) | Methylation in Control | 5.09E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon85 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg23110957) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:1.04E-02; Z-score:5.53E-01 | ||
|
Methylation in Case |
8.73E-01 (Median) | Methylation in Control | 8.56E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon86 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg00215425) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:1.25E-02; Z-score:3.92E-01 | ||
|
Methylation in Case |
8.91E-01 (Median) | Methylation in Control | 8.71E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon87 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg08516247) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:1.28E-02; Z-score:4.11E-01 | ||
|
Methylation in Case |
9.52E-01 (Median) | Methylation in Control | 9.44E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon88 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg17788850) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.30E-02; Z-score:-4.46E-01 | ||
|
Methylation in Case |
8.69E-01 (Median) | Methylation in Control | 8.75E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon89 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg10620395) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:1.35E-02; Z-score:8.38E-01 | ||
|
Methylation in Case |
8.27E-01 (Median) | Methylation in Control | 8.05E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon90 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg00509649) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:1.72E-02; Z-score:-7.33E-01 | ||
|
Methylation in Case |
1.38E-01 (Median) | Methylation in Control | 1.87E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon91 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg20422819) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.55E-02; Z-score:-2.48E-01 | ||
|
Methylation in Case |
7.27E-01 (Median) | Methylation in Control | 7.33E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon92 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg08344943) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:2.95E-02; Z-score:-1.54E-03 | ||
|
Methylation in Case |
9.46E-01 (Median) | Methylation in Control | 9.46E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon93 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg02762475) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.21E+00 | Statistic Test | p-value:3.00E-02; Z-score:-9.08E-01 | ||
|
Methylation in Case |
1.69E-01 (Median) | Methylation in Control | 2.05E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon94 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg02350636) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:3.58E-02; Z-score:5.10E-01 | ||
|
Methylation in Case |
9.35E-01 (Median) | Methylation in Control | 9.23E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon95 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg02039689) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:3.71E-02; Z-score:1.00E+00 | ||
|
Methylation in Case |
8.80E-01 (Median) | Methylation in Control | 8.63E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon96 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg22955555) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:4.22E-02; Z-score:4.87E-01 | ||
|
Methylation in Case |
8.20E-01 (Median) | Methylation in Control | 8.07E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon97 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg08351607) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.46E-02; Z-score:-5.65E-01 | ||
|
Methylation in Case |
8.15E-01 (Median) | Methylation in Control | 8.25E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon98 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg11805188) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:4.76E-02; Z-score:3.82E-01 | ||
|
Methylation in Case |
4.40E-01 (Median) | Methylation in Control | 4.14E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon99 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
Body (cg02079348) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:4.95E-02; Z-score:5.20E-01 | ||
|
Methylation in Case |
7.14E-01 (Median) | Methylation in Control | 6.98E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon100 |
Methylation of SLC12A7 in hepatocellular carcinoma | [ 8 ] | |||
|
Location |
3'UTR (cg07501827) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.20E+00 | Statistic Test | p-value:1.52E-09; Z-score:-2.08E+00 | ||
|
Methylation in Case |
5.13E-01 (Median) | Methylation in Control | 6.13E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
HIV infection |
58 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
TSS1500 (cg02739870) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.33E+00 | Statistic Test | p-value:4.41E-06; Z-score:1.79E+00 | ||
|
Methylation in Case |
4.42E-01 (Median) | Methylation in Control | 3.32E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
TSS1500 (cg24000908) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.24E+00 | Statistic Test | p-value:3.00E-05; Z-score:1.56E+00 | ||
|
Methylation in Case |
3.87E-01 (Median) | Methylation in Control | 3.13E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
TSS1500 (cg15083845) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:4.35E-02; Z-score:1.86E-01 | ||
|
Methylation in Case |
8.67E-01 (Median) | Methylation in Control | 8.62E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
TSS200 (cg23091824) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.24E+00 | Statistic Test | p-value:1.11E-02; Z-score:4.96E-01 | ||
|
Methylation in Case |
5.79E-02 (Median) | Methylation in Control | 4.67E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
TSS200 (cg21985251) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.32E+00 | Statistic Test | p-value:1.13E-02; Z-score:6.72E-01 | ||
|
Methylation in Case |
1.04E-01 (Median) | Methylation in Control | 7.92E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
TSS200 (cg15680989) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.30E+00 | Statistic Test | p-value:1.24E-02; Z-score:8.79E-01 | ||
|
Methylation in Case |
3.70E-02 (Median) | Methylation in Control | 2.85E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg22772691) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:2.33E-10; Z-score:1.85E+00 | ||
|
Methylation in Case |
4.65E-01 (Median) | Methylation in Control | 3.84E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg00509649) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.45E+00 | Statistic Test | p-value:1.39E-09; Z-score:1.67E+00 | ||
|
Methylation in Case |
3.59E-01 (Median) | Methylation in Control | 2.47E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg01171355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:3.07E-09; Z-score:1.08E+00 | ||
|
Methylation in Case |
9.63E-01 (Median) | Methylation in Control | 9.34E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg11235297) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.69E+00 | Statistic Test | p-value:5.47E-09; Z-score:3.30E+00 | ||
|
Methylation in Case |
5.87E-01 (Median) | Methylation in Control | 3.47E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg16017429) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:5.81E-09; Z-score:8.41E-01 | ||
|
Methylation in Case |
9.94E-01 (Median) | Methylation in Control | 9.85E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg20730619) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:1.27E-08; Z-score:1.21E+00 | ||
|
Methylation in Case |
8.75E-01 (Median) | Methylation in Control | 8.49E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg08543327) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.77E+00 | Statistic Test | p-value:1.83E-08; Z-score:3.02E+00 | ||
|
Methylation in Case |
4.16E-01 (Median) | Methylation in Control | 2.35E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg21946374) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.38E+00 | Statistic Test | p-value:2.33E-08; Z-score:3.02E+00 | ||
|
Methylation in Case |
5.57E-01 (Median) | Methylation in Control | 4.04E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg19773466) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:1.13E-07; Z-score:3.46E+00 | ||
|
Methylation in Case |
5.18E-01 (Median) | Methylation in Control | 4.32E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg18997983) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.42E+00 | Statistic Test | p-value:2.19E-07; Z-score:3.13E+00 | ||
|
Methylation in Case |
6.26E-01 (Median) | Methylation in Control | 4.41E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg02382320) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.00E+00 | Statistic Test | p-value:1.22E-06; Z-score:8.37E-01 | ||
|
Methylation in Case |
9.88E-01 (Median) | Methylation in Control | 9.83E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg13681701) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.18E+00 | Statistic Test | p-value:2.50E-06; Z-score:1.09E+00 | ||
|
Methylation in Case |
3.27E-01 (Median) | Methylation in Control | 2.78E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg10620395) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:2.93E-06; Z-score:1.32E+00 | ||
|
Methylation in Case |
8.01E-01 (Median) | Methylation in Control | 7.50E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg05819249) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:3.80E-06; Z-score:1.24E+00 | ||
|
Methylation in Case |
8.33E-01 (Median) | Methylation in Control | 7.61E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg11677852) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:7.40E-06; Z-score:2.03E+00 | ||
|
Methylation in Case |
7.17E-01 (Median) | Methylation in Control | 6.02E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg04380229) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:8.25E-06; Z-score:5.15E-01 | ||
|
Methylation in Case |
9.06E-01 (Median) | Methylation in Control | 8.86E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg08830157) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:1.32E-05; Z-score:1.11E+00 | ||
|
Methylation in Case |
3.37E-01 (Median) | Methylation in Control | 2.88E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg27072813) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:1.98E-05; Z-score:-1.67E+00 | ||
|
Methylation in Case |
6.75E-01 (Median) | Methylation in Control | 7.50E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg23989709) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:1.99E-05; Z-score:7.03E-01 | ||
|
Methylation in Case |
8.88E-01 (Median) | Methylation in Control | 8.73E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg12146829) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.54E+00 | Statistic Test | p-value:2.56E-05; Z-score:1.60E+00 | ||
|
Methylation in Case |
1.70E-01 (Median) | Methylation in Control | 1.11E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg02762475) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.48E+00 | Statistic Test | p-value:3.62E-05; Z-score:-1.41E+00 | ||
|
Methylation in Case |
2.16E-01 (Median) | Methylation in Control | 3.19E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg19854293) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:5.28E-05; Z-score:1.07E+00 | ||
|
Methylation in Case |
8.90E-01 (Median) | Methylation in Control | 8.63E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg13376768) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:7.51E-05; Z-score:8.07E-01 | ||
|
Methylation in Case |
9.70E-01 (Median) | Methylation in Control | 9.57E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg22172143) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.44E+00 | Statistic Test | p-value:2.27E-04; Z-score:1.45E+00 | ||
|
Methylation in Case |
2.25E-01 (Median) | Methylation in Control | 1.56E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg23503101) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:2.79E-04; Z-score:1.16E+00 | ||
|
Methylation in Case |
8.40E-01 (Median) | Methylation in Control | 8.07E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg16246240) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:2.90E-04; Z-score:-1.93E+00 | ||
|
Methylation in Case |
6.00E-01 (Median) | Methylation in Control | 6.88E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg24117063) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:3.47E-04; Z-score:1.02E+00 | ||
|
Methylation in Case |
9.28E-01 (Median) | Methylation in Control | 9.09E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg08516247) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:5.89E-04; Z-score:8.34E-01 | ||
|
Methylation in Case |
8.97E-01 (Median) | Methylation in Control | 8.76E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg16419756) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:9.49E-04; Z-score:-1.82E+00 | ||
|
Methylation in Case |
7.81E-01 (Median) | Methylation in Control | 8.36E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg14596589) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:1.39E-03; Z-score:5.92E-01 | ||
|
Methylation in Case |
6.27E-01 (Median) | Methylation in Control | 5.75E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon37 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg14817049) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:2.14E-03; Z-score:5.92E-01 | ||
|
Methylation in Case |
9.52E-01 (Median) | Methylation in Control | 9.40E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon38 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg18756954) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.00E+00 | Statistic Test | p-value:2.41E-03; Z-score:5.23E-01 | ||
|
Methylation in Case |
9.90E-01 (Median) | Methylation in Control | 9.88E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon39 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg10594510) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.72E-03; Z-score:-6.43E-01 | ||
|
Methylation in Case |
9.61E-01 (Median) | Methylation in Control | 9.68E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon40 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg00063291) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:4.01E-03; Z-score:7.45E-01 | ||
|
Methylation in Case |
8.76E-01 (Median) | Methylation in Control | 8.57E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon41 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg02079348) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:4.26E-03; Z-score:-9.05E-01 | ||
|
Methylation in Case |
7.43E-01 (Median) | Methylation in Control | 7.71E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon42 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg17851021) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:4.48E-03; Z-score:-1.82E+00 | ||
|
Methylation in Case |
8.20E-01 (Median) | Methylation in Control | 8.65E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon43 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg24886748) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.32E+00 | Statistic Test | p-value:4.87E-03; Z-score:7.67E-01 | ||
|
Methylation in Case |
3.27E-02 (Median) | Methylation in Control | 2.48E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon44 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg24059022) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:6.03E-03; Z-score:7.78E-01 | ||
|
Methylation in Case |
9.53E-01 (Median) | Methylation in Control | 9.41E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon45 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg05978154) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:6.11E-03; Z-score:-3.39E-01 | ||
|
Methylation in Case |
9.81E-01 (Median) | Methylation in Control | 9.84E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon46 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg13299707) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:8.11E-03; Z-score:3.76E-01 | ||
|
Methylation in Case |
7.90E-01 (Median) | Methylation in Control | 7.79E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon47 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg05163933) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.08E-02; Z-score:-1.28E+00 | ||
|
Methylation in Case |
7.09E-01 (Median) | Methylation in Control | 7.52E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon48 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg23110957) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.21E-02; Z-score:-4.12E-01 | ||
|
Methylation in Case |
8.95E-01 (Median) | Methylation in Control | 9.06E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon49 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg06973176) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:1.75E-02; Z-score:5.09E-01 | ||
|
Methylation in Case |
8.62E-01 (Median) | Methylation in Control | 8.52E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon50 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg23115083) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.76E-02; Z-score:-8.31E-01 | ||
|
Methylation in Case |
8.46E-01 (Median) | Methylation in Control | 8.67E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon51 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg03281154) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.85E-02; Z-score:-9.81E-01 | ||
|
Methylation in Case |
8.29E-01 (Median) | Methylation in Control | 8.55E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon52 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg04114636) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:1.87E-02; Z-score:1.02E+00 | ||
|
Methylation in Case |
8.80E-01 (Median) | Methylation in Control | 8.60E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon53 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg18677871) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:2.08E-02; Z-score:8.32E-01 | ||
|
Methylation in Case |
8.41E-01 (Median) | Methylation in Control | 8.14E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon54 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg14047153) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:2.33E-02; Z-score:7.09E-01 | ||
|
Methylation in Case |
9.89E-01 (Median) | Methylation in Control | 9.83E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon55 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg06344195) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:3.34E-02; Z-score:5.11E-01 | ||
|
Methylation in Case |
9.14E-01 (Median) | Methylation in Control | 9.03E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon56 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg16044603) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:3.72E-02; Z-score:7.54E-01 | ||
|
Methylation in Case |
8.70E-01 (Median) | Methylation in Control | 8.47E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon57 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
Body (cg15601915) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:4.53E-02; Z-score:5.95E-01 | ||
|
Methylation in Case |
7.85E-01 (Median) | Methylation in Control | 7.69E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon58 |
Methylation of SLC12A7 in HIV infection | [ 9 ] | |||
|
Location |
3'UTR (cg17568547) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:3.14E-03; Z-score:5.38E-01 | ||
|
Methylation in Case |
9.05E-01 (Median) | Methylation in Control | 8.96E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
48 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
TSS1500 (cg02739870) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.32E+00 | Statistic Test | p-value:1.55E-03; Z-score:-1.93E+00 | ||
|
Methylation in Case |
5.60E-01 (Median) | Methylation in Control | 7.38E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
TSS1500 (cg24000908) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.29E+00 | Statistic Test | p-value:1.84E-03; Z-score:-2.01E+00 | ||
|
Methylation in Case |
5.50E-01 (Median) | Methylation in Control | 7.10E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg18677871) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.18E+00 | Statistic Test | p-value:1.23E-04; Z-score:2.20E+00 | ||
|
Methylation in Case |
6.99E-01 (Median) | Methylation in Control | 5.90E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg01266985) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:1.93E-04; Z-score:2.11E+00 | ||
|
Methylation in Case |
7.64E-01 (Median) | Methylation in Control | 6.75E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg00509649) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.87E+00 | Statistic Test | p-value:2.77E-04; Z-score:5.44E+00 | ||
|
Methylation in Case |
4.61E-01 (Median) | Methylation in Control | 2.46E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg23221540) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:3.62E-04; Z-score:1.96E+00 | ||
|
Methylation in Case |
8.65E-01 (Median) | Methylation in Control | 7.93E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg13681701) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.64E+00 | Statistic Test | p-value:3.66E-04; Z-score:4.74E+00 | ||
|
Methylation in Case |
4.48E-01 (Median) | Methylation in Control | 2.72E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg21123417) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.42E+00 | Statistic Test | p-value:4.26E-04; Z-score:2.22E+00 | ||
|
Methylation in Case |
7.04E-01 (Median) | Methylation in Control | 4.95E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg10620395) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:5.94E-04; Z-score:-1.93E+00 | ||
|
Methylation in Case |
8.07E-01 (Median) | Methylation in Control | 8.36E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg26500588) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:7.55E-04; Z-score:-2.39E+00 | ||
|
Methylation in Case |
7.72E-01 (Median) | Methylation in Control | 8.19E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg00063291) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:8.66E-04; Z-score:-1.94E+00 | ||
|
Methylation in Case |
8.54E-01 (Median) | Methylation in Control | 8.93E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg26213438) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.14E-03; Z-score:-2.99E+00 | ||
|
Methylation in Case |
7.90E-01 (Median) | Methylation in Control | 8.35E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg13299707) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.34E-03; Z-score:-1.67E+00 | ||
|
Methylation in Case |
7.90E-01 (Median) | Methylation in Control | 8.20E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg21513991) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.90E-03; Z-score:-2.32E+00 | ||
|
Methylation in Case |
8.14E-01 (Median) | Methylation in Control | 8.65E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg02079348) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.28E-03; Z-score:-1.56E+00 | ||
|
Methylation in Case |
7.29E-01 (Median) | Methylation in Control | 7.68E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg00697639) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:3.32E-03; Z-score:-2.26E+00 | ||
|
Methylation in Case |
8.27E-01 (Median) | Methylation in Control | 8.61E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg16997104) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:4.10E-03; Z-score:-1.66E+00 | ||
|
Methylation in Case |
9.18E-01 (Median) | Methylation in Control | 9.44E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg06637017) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:5.25E-03; Z-score:1.21E+00 | ||
|
Methylation in Case |
8.21E-01 (Median) | Methylation in Control | 7.62E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg07459121) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:6.57E-03; Z-score:2.18E+00 | ||
|
Methylation in Case |
8.54E-01 (Median) | Methylation in Control | 7.29E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg02350636) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:7.28E-03; Z-score:-1.88E+00 | ||
|
Methylation in Case |
9.14E-01 (Median) | Methylation in Control | 9.39E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg22772691) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:8.67E-03; Z-score:2.75E+00 | ||
|
Methylation in Case |
5.50E-01 (Median) | Methylation in Control | 4.56E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg18576686) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.06E-02; Z-score:-1.35E+00 | ||
|
Methylation in Case |
8.07E-01 (Median) | Methylation in Control | 8.35E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg17851021) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.13E-02; Z-score:-1.77E+00 | ||
|
Methylation in Case |
8.59E-01 (Median) | Methylation in Control | 8.81E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg22955595) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.47E-02; Z-score:-1.39E+00 | ||
|
Methylation in Case |
7.85E-01 (Median) | Methylation in Control | 8.14E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg23115083) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:1.50E-02; Z-score:-1.43E+00 | ||
|
Methylation in Case |
8.31E-01 (Median) | Methylation in Control | 8.70E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg03281154) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.62E-02; Z-score:-1.63E+00 | ||
|
Methylation in Case |
7.84E-01 (Median) | Methylation in Control | 8.18E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg11713480) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.86E-02; Z-score:-1.09E+00 | ||
|
Methylation in Case |
8.74E-01 (Median) | Methylation in Control | 8.91E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg10857489) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.88E-02; Z-score:-1.40E+00 | ||
|
Methylation in Case |
8.98E-01 (Median) | Methylation in Control | 9.12E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg08702805) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.98E-02; Z-score:-9.86E-01 | ||
|
Methylation in Case |
7.90E-01 (Median) | Methylation in Control | 8.13E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg27072813) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:2.69E-02; Z-score:-1.70E+00 | ||
|
Methylation in Case |
7.56E-01 (Median) | Methylation in Control | 8.01E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg21516614) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.80E-02; Z-score:-1.33E+00 | ||
|
Methylation in Case |
9.58E-01 (Median) | Methylation in Control | 9.66E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg14671982) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:2.83E-02; Z-score:-1.23E+00 | ||
|
Methylation in Case |
7.73E-01 (Median) | Methylation in Control | 8.07E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg16017429) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.89E-02; Z-score:-2.50E+00 | ||
|
Methylation in Case |
9.59E-01 (Median) | Methylation in Control | 9.71E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg11805188) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:3.17E-02; Z-score:1.32E+00 | ||
|
Methylation in Case |
7.29E-01 (Median) | Methylation in Control | 6.66E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg12993807) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.26E-02; Z-score:-4.18E+00 | ||
|
Methylation in Case |
9.57E-01 (Median) | Methylation in Control | 9.67E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg26439015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.59E+00 | Statistic Test | p-value:3.26E-02; Z-score:1.97E+00 | ||
|
Methylation in Case |
3.56E-02 (Median) | Methylation in Control | 2.24E-02 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon37 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg15647725) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:3.28E-02; Z-score:1.02E+00 | ||
|
Methylation in Case |
6.60E-01 (Median) | Methylation in Control | 6.09E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon38 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg16246240) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:3.38E-02; Z-score:-8.76E-01 | ||
|
Methylation in Case |
6.60E-01 (Median) | Methylation in Control | 6.89E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon39 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg20730619) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:3.47E-02; Z-score:-1.43E+00 | ||
|
Methylation in Case |
8.46E-01 (Median) | Methylation in Control | 8.69E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon40 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg14047153) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:4.10E-02; Z-score:-2.07E+00 | ||
|
Methylation in Case |
9.57E-01 (Median) | Methylation in Control | 9.71E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon41 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg00601711) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:4.19E-02; Z-score:8.15E-01 | ||
|
Methylation in Case |
8.10E-01 (Median) | Methylation in Control | 7.84E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon42 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg26824947) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:4.24E-02; Z-score:-2.02E+00 | ||
|
Methylation in Case |
7.05E-01 (Median) | Methylation in Control | 7.44E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon43 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg06058262) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:4.46E-02; Z-score:-1.51E+00 | ||
|
Methylation in Case |
8.84E-01 (Median) | Methylation in Control | 9.00E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon44 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg08830157) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:4.53E-02; Z-score:-1.55E+00 | ||
|
Methylation in Case |
5.72E-01 (Median) | Methylation in Control | 6.17E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon45 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg17969789) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:4.76E-02; Z-score:-7.66E-01 | ||
|
Methylation in Case |
7.83E-01 (Median) | Methylation in Control | 8.00E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon46 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg18196463) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.78E-02; Z-score:-1.68E+00 | ||
|
Methylation in Case |
9.57E-01 (Median) | Methylation in Control | 9.66E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon47 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
Body (cg23998435) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.96E-02; Z-score:-7.75E-01 | ||
|
Methylation in Case |
9.44E-01 (Median) | Methylation in Control | 9.49E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon48 |
Methylation of SLC12A7 in lung adenocarcinoma | [ 10 ] | |||
|
Location |
3'UTR (cg19086001) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.60E-02; Z-score:-1.04E+00 | ||
|
Methylation in Case |
8.16E-01 (Median) | Methylation in Control | 8.36E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
52 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
TSS1500 (cg02739870) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:2.46E-10; Z-score:-1.62E+00 | ||
|
Methylation in Case |
7.58E-01 (Median) | Methylation in Control | 8.66E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
TSS1500 (cg24000908) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:3.94E-05; Z-score:-9.74E-01 | ||
|
Methylation in Case |
8.16E-01 (Median) | Methylation in Control | 8.76E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg00601711) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:2.34E-09; Z-score:1.37E+00 | ||
|
Methylation in Case |
8.78E-01 (Median) | Methylation in Control | 8.23E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg23514135) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.73E-08; Z-score:-2.11E+00 | ||
|
Methylation in Case |
9.16E-01 (Median) | Methylation in Control | 9.50E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg02079348) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:1.61E-07; Z-score:-1.18E+00 | ||
|
Methylation in Case |
7.89E-01 (Median) | Methylation in Control | 8.30E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg26244838) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:3.88E-07; Z-score:-1.78E+00 | ||
|
Methylation in Case |
7.92E-01 (Median) | Methylation in Control | 8.58E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg13301368) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:4.40E-07; Z-score:1.15E+00 | ||
|
Methylation in Case |
9.15E-01 (Median) | Methylation in Control | 8.86E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg14671982) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:5.66E-07; Z-score:1.30E+00 | ||
|
Methylation in Case |
8.55E-01 (Median) | Methylation in Control | 8.14E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg21334510) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:6.24E-06; Z-score:8.95E-01 | ||
|
Methylation in Case |
6.20E-01 (Median) | Methylation in Control | 5.70E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg21946374) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:8.43E-06; Z-score:-7.03E-01 | ||
|
Methylation in Case |
8.15E-01 (Median) | Methylation in Control | 8.39E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg18677871) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:8.77E-06; Z-score:1.26E+00 | ||
|
Methylation in Case |
7.39E-01 (Median) | Methylation in Control | 6.81E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg00420510) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:1.80E-04; Z-score:1.39E+00 | ||
|
Methylation in Case |
8.23E-01 (Median) | Methylation in Control | 7.69E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg04380229) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:2.52E-04; Z-score:-7.83E-01 | ||
|
Methylation in Case |
8.75E-01 (Median) | Methylation in Control | 8.95E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg09547427) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:2.60E-04; Z-score:-8.52E-01 | ||
|
Methylation in Case |
8.78E-01 (Median) | Methylation in Control | 8.95E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg21513991) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.13E-04; Z-score:-6.58E-01 | ||
|
Methylation in Case |
9.15E-01 (Median) | Methylation in Control | 9.26E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg26213438) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:3.60E-04; Z-score:8.48E-01 | ||
|
Methylation in Case |
8.42E-01 (Median) | Methylation in Control | 8.16E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg00600029) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:5.09E-04; Z-score:-5.75E-01 | ||
|
Methylation in Case |
9.41E-01 (Median) | Methylation in Control | 9.46E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg04213775) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:5.80E-04; Z-score:-5.88E-01 | ||
|
Methylation in Case |
8.84E-01 (Median) | Methylation in Control | 8.95E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg16246240) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:7.37E-04; Z-score:8.47E-01 | ||
|
Methylation in Case |
7.25E-01 (Median) | Methylation in Control | 6.73E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg06344195) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:7.68E-04; Z-score:-7.56E-01 | ||
|
Methylation in Case |
9.35E-01 (Median) | Methylation in Control | 9.46E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg15601915) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:8.87E-04; Z-score:-7.49E-01 | ||
|
Methylation in Case |
8.36E-01 (Median) | Methylation in Control | 8.58E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg10620395) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:9.45E-04; Z-score:-7.41E-01 | ||
|
Methylation in Case |
9.15E-01 (Median) | Methylation in Control | 9.27E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg04114636) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.06E-03; Z-score:-6.12E-01 | ||
|
Methylation in Case |
9.01E-01 (Median) | Methylation in Control | 9.11E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg18655438) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.28E-03; Z-score:-7.15E-01 | ||
|
Methylation in Case |
8.91E-01 (Median) | Methylation in Control | 9.05E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg00346376) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.37E-03; Z-score:-5.74E-01 | ||
|
Methylation in Case |
8.23E-01 (Median) | Methylation in Control | 8.40E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg02382320) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:2.08E-03; Z-score:6.89E-01 | ||
|
Methylation in Case |
8.99E-01 (Median) | Methylation in Control | 8.85E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg05636015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.39E-03; Z-score:-7.08E-01 | ||
|
Methylation in Case |
8.96E-01 (Median) | Methylation in Control | 9.09E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg16163535) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.95E-03; Z-score:-5.56E-01 | ||
|
Methylation in Case |
9.43E-01 (Median) | Methylation in Control | 9.51E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg17733824) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:4.01E-03; Z-score:5.73E-01 | ||
|
Methylation in Case |
8.62E-01 (Median) | Methylation in Control | 8.46E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg26500588) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:4.05E-03; Z-score:-5.38E-01 | ||
|
Methylation in Case |
8.73E-01 (Median) | Methylation in Control | 8.88E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg23115083) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.09E-03; Z-score:-6.02E-01 | ||
|
Methylation in Case |
9.12E-01 (Median) | Methylation in Control | 9.25E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg01551729) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:7.66E-03; Z-score:5.82E-01 | ||
|
Methylation in Case |
8.58E-01 (Median) | Methylation in Control | 8.44E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg23491790) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:7.67E-03; Z-score:6.09E-01 | ||
|
Methylation in Case |
8.77E-01 (Median) | Methylation in Control | 8.57E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg00551954) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:8.57E-03; Z-score:-4.37E-01 | ||
|
Methylation in Case |
8.98E-01 (Median) | Methylation in Control | 9.06E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg06058262) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:9.50E-03; Z-score:-7.93E-01 | ||
|
Methylation in Case |
9.40E-01 (Median) | Methylation in Control | 9.48E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg02349468) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:9.93E-03; Z-score:-6.74E-01 | ||
|
Methylation in Case |
8.88E-01 (Median) | Methylation in Control | 9.06E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon37 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg25528709) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.12E-02; Z-score:-4.53E-01 | ||
|
Methylation in Case |
9.61E-01 (Median) | Methylation in Control | 9.64E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon38 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg00095276) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.48E-02; Z-score:-4.62E-01 | ||
|
Methylation in Case |
8.26E-01 (Median) | Methylation in Control | 8.41E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon39 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg08293408) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.54E-02; Z-score:-6.84E-01 | ||
|
Methylation in Case |
9.14E-01 (Median) | Methylation in Control | 9.26E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon40 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg25945676) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:1.83E-02; Z-score:4.48E-01 | ||
|
Methylation in Case |
8.37E-01 (Median) | Methylation in Control | 8.13E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon41 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg24928433) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.00E+00 | Statistic Test | p-value:2.31E-02; Z-score:2.30E-01 | ||
|
Methylation in Case |
9.48E-01 (Median) | Methylation in Control | 9.44E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon42 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg11235297) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:2.41E-02; Z-score:-4.39E-01 | ||
|
Methylation in Case |
8.70E-01 (Median) | Methylation in Control | 8.89E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon43 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg25970471) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.44E-02; Z-score:-4.66E-01 | ||
|
Methylation in Case |
8.38E-01 (Median) | Methylation in Control | 8.50E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon44 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg21123417) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:2.64E-02; Z-score:8.29E-01 | ||
|
Methylation in Case |
7.02E-01 (Median) | Methylation in Control | 6.52E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon45 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg16997104) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:2.76E-02; Z-score:6.04E-01 | ||
|
Methylation in Case |
8.95E-01 (Median) | Methylation in Control | 8.77E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon46 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg11628781) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:2.79E-02; Z-score:-3.74E-01 | ||
|
Methylation in Case |
9.40E-01 (Median) | Methylation in Control | 9.44E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon47 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg09951201) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:2.80E-02; Z-score:4.56E-01 | ||
|
Methylation in Case |
8.86E-01 (Median) | Methylation in Control | 8.63E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon48 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg12199905) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.67E-02; Z-score:-5.99E-01 | ||
|
Methylation in Case |
9.38E-01 (Median) | Methylation in Control | 9.46E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon49 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg00063291) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.85E-02; Z-score:-3.47E-01 | ||
|
Methylation in Case |
9.47E-01 (Median) | Methylation in Control | 9.53E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon50 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg00098175) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.02E-02; Z-score:-3.92E-01 | ||
|
Methylation in Case |
8.85E-01 (Median) | Methylation in Control | 8.92E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon51 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg15597069) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.12E-02; Z-score:-4.88E-01 | ||
|
Methylation in Case |
9.46E-01 (Median) | Methylation in Control | 9.52E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon52 |
Methylation of SLC12A7 in papillary thyroid cancer | [ 11 ] | |||
|
Location |
Body (cg08351607) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.48E-02; Z-score:-3.57E-01 | ||
|
Methylation in Case |
8.82E-01 (Median) | Methylation in Control | 8.88E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Systemic lupus erythematosus |
20 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
TSS200 (cg21985251) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:3.33E-02; Z-score:-3.75E-02 | ||
|
Methylation in Case |
4.24E-02 (Median) | Methylation in Control | 4.33E-02 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg25945676) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.02E-05; Z-score:-1.52E-01 | ||
|
Methylation in Case |
9.20E-01 (Median) | Methylation in Control | 9.23E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg11422312) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.82E-03; Z-score:-4.56E-02 | ||
|
Methylation in Case |
8.87E-01 (Median) | Methylation in Control | 8.92E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg26439015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:2.82E-03; Z-score:-8.71E-02 | ||
|
Methylation in Case |
9.39E-03 (Median) | Methylation in Control | 1.02E-02 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg02295574) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:7.81E-03; Z-score:-1.04E-01 | ||
|
Methylation in Case |
8.86E-01 (Median) | Methylation in Control | 8.91E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg04380229) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:8.32E-03; Z-score:-2.61E-01 | ||
|
Methylation in Case |
8.90E-01 (Median) | Methylation in Control | 8.98E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg21123417) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:8.88E-03; Z-score:-2.83E-01 | ||
|
Methylation in Case |
8.74E-01 (Median) | Methylation in Control | 8.82E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg14981610) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:9.26E-03; Z-score:-7.91E-02 | ||
|
Methylation in Case |
9.26E-01 (Median) | Methylation in Control | 9.27E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg17097710) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:9.68E-03; Z-score:-2.08E-01 | ||
|
Methylation in Case |
9.24E-01 (Median) | Methylation in Control | 9.27E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg13301368) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.59E-02; Z-score:-2.48E-01 | ||
|
Methylation in Case |
9.29E-01 (Median) | Methylation in Control | 9.32E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg25404678) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.79E-02; Z-score:-1.91E-01 | ||
|
Methylation in Case |
9.78E-01 (Median) | Methylation in Control | 9.79E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg08516247) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:2.16E-02; Z-score:-1.44E-01 | ||
|
Methylation in Case |
9.22E-01 (Median) | Methylation in Control | 9.25E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg23948452) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:2.26E-02; Z-score:-2.92E-01 | ||
|
Methylation in Case |
9.36E-01 (Median) | Methylation in Control | 9.41E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg13592947) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.68E-02; Z-score:-2.82E-01 | ||
|
Methylation in Case |
4.72E-01 (Median) | Methylation in Control | 4.97E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg04114636) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:3.15E-02; Z-score:-5.95E-02 | ||
|
Methylation in Case |
8.96E-01 (Median) | Methylation in Control | 8.97E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg23491790) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:3.43E-02; Z-score:-7.52E-02 | ||
|
Methylation in Case |
9.62E-01 (Median) | Methylation in Control | 9.63E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg16419756) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.90E-02; Z-score:-2.05E-01 | ||
|
Methylation in Case |
8.50E-01 (Median) | Methylation in Control | 8.57E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg11628781) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:4.29E-02; Z-score:-1.88E-01 | ||
|
Methylation in Case |
9.04E-01 (Median) | Methylation in Control | 9.08E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg00509649) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:4.47E-02; Z-score:2.37E-01 | ||
|
Methylation in Case |
3.33E-01 (Median) | Methylation in Control | 2.98E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC12A7 in systemic lupus erythematosus | [ 12 ] | |||
|
Location |
Body (cg11713480) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:4.74E-02; Z-score:-1.36E-01 | ||
|
Methylation in Case |
9.15E-01 (Median) | Methylation in Control | 9.17E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Atypical teratoid rhabdoid tumor |
86 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg00063291) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.25E+00 | Statistic Test | p-value:1.59E-05; Z-score:1.11E+00 | ||
|
Methylation in Case |
7.11E-01 (Median) | Methylation in Control | 5.67E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg00095276) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:1.69E-05; Z-score:1.12E+00 | ||
|
Methylation in Case |
8.91E-01 (Median) | Methylation in Control | 8.26E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg00098175) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:1.71E-05; Z-score:7.88E-01 | ||
|
Methylation in Case |
9.01E-01 (Median) | Methylation in Control | 8.27E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg00215425) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:1.94E-05; Z-score:6.76E-01 | ||
|
Methylation in Case |
8.74E-01 (Median) | Methylation in Control | 7.45E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg00278107) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:1.98E-05; Z-score:8.70E-01 | ||
|
Methylation in Case |
9.01E-01 (Median) | Methylation in Control | 7.82E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg00346376) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.16E+00 | Statistic Test | p-value:2.18E-05; Z-score:1.18E+00 | ||
|
Methylation in Case |
8.79E-01 (Median) | Methylation in Control | 7.57E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg00420510) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:2.48E-05; Z-score:-1.00E+00 | ||
|
Methylation in Case |
8.42E-01 (Median) | Methylation in Control | 8.88E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg00509649) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:2.82E-05; Z-score:1.16E+00 | ||
|
Methylation in Case |
9.38E-01 (Median) | Methylation in Control | 9.14E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg00551954) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:2.98E-05; Z-score:6.43E-01 | ||
|
Methylation in Case |
8.92E-01 (Median) | Methylation in Control | 7.65E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg00600029) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.39E+00 | Statistic Test | p-value:3.02E-05; Z-score:1.17E+00 | ||
|
Methylation in Case |
9.55E-01 (Median) | Methylation in Control | 6.89E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg00601711) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:3.03E-05; Z-score:-1.12E+00 | ||
|
Methylation in Case |
8.11E-01 (Median) | Methylation in Control | 8.65E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg00697639) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:3.32E-05; Z-score:1.10E+00 | ||
|
Methylation in Case |
7.99E-01 (Median) | Methylation in Control | 6.68E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg01171355) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:5.24E-05; Z-score:-1.01E+00 | ||
|
Methylation in Case |
4.58E-01 (Median) | Methylation in Control | 5.60E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg01266985) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:5.73E-05; Z-score:-4.98E-01 | ||
|
Methylation in Case |
7.49E-01 (Median) | Methylation in Control | 7.90E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg01551729) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.81E+00 | Statistic Test | p-value:7.38E-05; Z-score:-1.15E+00 | ||
|
Methylation in Case |
2.23E-01 (Median) | Methylation in Control | 4.03E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg01903305) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.68E+00 | Statistic Test | p-value:9.22E-05; Z-score:8.22E-01 | ||
|
Methylation in Case |
1.57E-01 (Median) | Methylation in Control | 9.32E-02 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg02027730) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:1.08E-04; Z-score:-5.51E-01 | ||
|
Methylation in Case |
7.71E-01 (Median) | Methylation in Control | 8.09E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg02039689) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:1.11E-04; Z-score:1.20E+00 | ||
|
Methylation in Case |
7.31E-01 (Median) | Methylation in Control | 6.06E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon19 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg02079348) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:1.12E-04; Z-score:1.03E+00 | ||
|
Methylation in Case |
8.80E-01 (Median) | Methylation in Control | 8.03E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon20 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg02295574) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.23E+00 | Statistic Test | p-value:1.42E-04; Z-score:-6.30E-01 | ||
|
Methylation in Case |
4.32E-01 (Median) | Methylation in Control | 5.31E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon21 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg02333352) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:1.45E-04; Z-score:-8.71E-01 | ||
|
Methylation in Case |
4.30E-01 (Median) | Methylation in Control | 5.26E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon22 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg02349468) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:1.53E-04; Z-score:-1.03E+00 | ||
|
Methylation in Case |
6.90E-01 (Median) | Methylation in Control | 7.67E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon23 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg02350636) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.55E-04; Z-score:-5.87E-01 | ||
|
Methylation in Case |
8.23E-01 (Median) | Methylation in Control | 8.76E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon24 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg02382320) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:1.61E-04; Z-score:1.13E+00 | ||
|
Methylation in Case |
9.21E-01 (Median) | Methylation in Control | 8.63E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon25 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg02557110) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:1.76E-04; Z-score:9.84E-01 | ||
|
Methylation in Case |
8.69E-01 (Median) | Methylation in Control | 7.79E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon26 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg02762475) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:2.27E-04; Z-score:-8.26E-01 | ||
|
Methylation in Case |
8.65E-01 (Median) | Methylation in Control | 8.98E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon27 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg03281154) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:2.72E-04; Z-score:1.12E+00 | ||
|
Methylation in Case |
8.11E-01 (Median) | Methylation in Control | 7.44E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon28 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg03343453) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:2.91E-04; Z-score:1.16E+00 | ||
|
Methylation in Case |
8.87E-01 (Median) | Methylation in Control | 7.33E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon29 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg04114636) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:5.57E-04; Z-score:5.70E-01 | ||
|
Methylation in Case |
8.93E-01 (Median) | Methylation in Control | 8.33E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon30 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg04184427) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:5.98E-04; Z-score:-8.65E-01 | ||
|
Methylation in Case |
8.42E-01 (Median) | Methylation in Control | 8.91E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon31 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg04213775) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:6.22E-04; Z-score:-5.70E-01 | ||
|
Methylation in Case |
9.34E-01 (Median) | Methylation in Control | 9.52E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon32 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg04226648) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:6.31E-04; Z-score:-6.24E-01 | ||
|
Methylation in Case |
8.66E-01 (Median) | Methylation in Control | 8.92E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon33 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg04380229) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:7.38E-04; Z-score:6.52E-01 | ||
|
Methylation in Case |
9.18E-01 (Median) | Methylation in Control | 8.45E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon34 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg04467119) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.29E+00 | Statistic Test | p-value:7.87E-04; Z-score:9.96E-01 | ||
|
Methylation in Case |
4.22E-01 (Median) | Methylation in Control | 3.27E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon35 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg04725215) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.53E+00 | Statistic Test | p-value:9.37E-04; Z-score:-1.05E+00 | ||
|
Methylation in Case |
2.00E-01 (Median) | Methylation in Control | 3.07E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon36 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg05163933) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.19E+00 | Statistic Test | p-value:1.30E-03; Z-score:-6.86E-01 | ||
|
Methylation in Case |
2.46E-01 (Median) | Methylation in Control | 2.94E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon37 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg05636015) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:1.58E-03; Z-score:6.13E-01 | ||
|
Methylation in Case |
8.20E-01 (Median) | Methylation in Control | 7.55E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon38 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg05819249) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.21E+00 | Statistic Test | p-value:1.68E-03; Z-score:-5.61E-01 | ||
|
Methylation in Case |
2.02E-01 (Median) | Methylation in Control | 2.45E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon39 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg05972654) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.64E+00 | Statistic Test | p-value:2.00E-03; Z-score:-5.11E-01 | ||
|
Methylation in Case |
7.60E-02 (Median) | Methylation in Control | 1.25E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon40 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg05978154) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.32E+00 | Statistic Test | p-value:2.02E-03; Z-score:1.08E+00 | ||
|
Methylation in Case |
4.60E-01 (Median) | Methylation in Control | 3.50E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon41 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg06058262) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:2.15E-03; Z-score:5.04E-01 | ||
|
Methylation in Case |
6.47E-01 (Median) | Methylation in Control | 6.03E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon42 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg06344195) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:2.38E-03; Z-score:-1.04E+00 | ||
|
Methylation in Case |
6.46E-01 (Median) | Methylation in Control | 7.88E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon43 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg06407917) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:2.45E-03; Z-score:1.65E-01 | ||
|
Methylation in Case |
7.79E-02 (Median) | Methylation in Control | 7.61E-02 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon44 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg06592065) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.21E+00 | Statistic Test | p-value:2.64E-03; Z-score:7.14E-01 | ||
|
Methylation in Case |
6.91E-01 (Median) | Methylation in Control | 5.71E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon45 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg06637017) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:2.71E-03; Z-score:4.18E-01 | ||
|
Methylation in Case |
9.62E-02 (Median) | Methylation in Control | 8.67E-02 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon46 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg06973176) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:3.17E-03; Z-score:6.55E-01 | ||
|
Methylation in Case |
8.98E-01 (Median) | Methylation in Control | 8.38E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon47 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg07459121) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:3.94E-03; Z-score:-8.01E-01 | ||
|
Methylation in Case |
7.24E-01 (Median) | Methylation in Control | 7.86E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon48 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg07567497) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.69E+00 | Statistic Test | p-value:4.43E-03; Z-score:-6.18E-01 | ||
|
Methylation in Case |
4.08E-02 (Median) | Methylation in Control | 1.10E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon49 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg08293408) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:5.70E-03; Z-score:3.29E-01 | ||
|
Methylation in Case |
8.18E-01 (Median) | Methylation in Control | 7.83E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon50 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg08344943) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:5.83E-03; Z-score:-3.07E-01 | ||
|
Methylation in Case |
9.23E-01 (Median) | Methylation in Control | 9.34E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon51 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg08351607) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.30E+00 | Statistic Test | p-value:5.89E-03; Z-score:8.31E-01 | ||
|
Methylation in Case |
5.91E-01 (Median) | Methylation in Control | 4.54E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon52 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg08382946) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:6.08E-03; Z-score:-7.23E-01 | ||
|
Methylation in Case |
4.57E-01 (Median) | Methylation in Control | 5.06E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon53 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg08516247) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:7.22E-03; Z-score:5.03E-01 | ||
|
Methylation in Case |
9.21E-01 (Median) | Methylation in Control | 8.97E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon54 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg08543327) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.23E+00 | Statistic Test | p-value:7.31E-03; Z-score:-7.02E-01 | ||
|
Methylation in Case |
4.21E-01 (Median) | Methylation in Control | 5.17E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon55 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg08702805) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.16E+00 | Statistic Test | p-value:7.72E-03; Z-score:5.19E-01 | ||
|
Methylation in Case |
4.92E-01 (Median) | Methylation in Control | 4.23E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon56 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg08830157) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.37E+00 | Statistic Test | p-value:8.03E-03; Z-score:-5.92E-01 | ||
|
Methylation in Case |
5.02E-02 (Median) | Methylation in Control | 1.19E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon57 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg09050160) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:8.90E-03; Z-score:2.76E-01 | ||
|
Methylation in Case |
2.47E-02 (Median) | Methylation in Control | 2.30E-02 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon58 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg09547427) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.10E-02; Z-score:-3.64E-01 | ||
|
Methylation in Case |
9.13E-01 (Median) | Methylation in Control | 9.25E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon59 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg09790628) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:1.19E-02; Z-score:5.69E-01 | ||
|
Methylation in Case |
5.42E-01 (Median) | Methylation in Control | 4.44E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon60 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg09951201) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:1.25E-02; Z-score:5.15E-01 | ||
|
Methylation in Case |
8.44E-01 (Median) | Methylation in Control | 7.96E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon61 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg10489284) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.65E+00 | Statistic Test | p-value:1.49E-02; Z-score:6.92E-01 | ||
|
Methylation in Case |
2.93E-01 (Median) | Methylation in Control | 1.78E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon62 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg10594510) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.62E-02; Z-score:-4.08E-01 | ||
|
Methylation in Case |
8.77E-01 (Median) | Methylation in Control | 9.01E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon63 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg10601043) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:1.63E-02; Z-score:3.33E-01 | ||
|
Methylation in Case |
8.94E-01 (Median) | Methylation in Control | 8.62E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon64 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg10620395) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:1.66E-02; Z-score:5.58E-01 | ||
|
Methylation in Case |
8.24E-01 (Median) | Methylation in Control | 7.67E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon65 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg10857489) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.94E+00 | Statistic Test | p-value:1.85E-02; Z-score:-8.21E-01 | ||
|
Methylation in Case |
5.66E-02 (Median) | Methylation in Control | 1.10E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon66 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg11235297) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:2.08E-02; Z-score:1.94E-01 | ||
|
Methylation in Case |
5.41E-02 (Median) | Methylation in Control | 5.05E-02 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon67 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg11422312) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.34E+00 | Statistic Test | p-value:2.22E-02; Z-score:-6.26E-01 | ||
|
Methylation in Case |
8.92E-02 (Median) | Methylation in Control | 1.20E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon68 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg11577329) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:2.32E-02; Z-score:3.07E-01 | ||
|
Methylation in Case |
6.19E-02 (Median) | Methylation in Control | 5.77E-02 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon69 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg11628781) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.51E+00 | Statistic Test | p-value:2.38E-02; Z-score:-1.04E+00 | ||
|
Methylation in Case |
1.21E-01 (Median) | Methylation in Control | 1.84E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon70 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg11677852) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:2.42E-02; Z-score:-1.67E-01 | ||
|
Methylation in Case |
9.35E-01 (Median) | Methylation in Control | 9.38E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon71 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg11713480) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:2.48E-02; Z-score:5.34E-01 | ||
|
Methylation in Case |
8.38E-01 (Median) | Methylation in Control | 8.14E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon72 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg11805188) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:2.50E-02; Z-score:6.03E-01 | ||
|
Methylation in Case |
8.98E-01 (Median) | Methylation in Control | 8.72E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon73 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg12146829) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:2.67E-02; Z-score:2.68E-01 | ||
|
Methylation in Case |
9.33E-01 (Median) | Methylation in Control | 9.22E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon74 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg12199905) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:2.70E-02; Z-score:3.12E-01 | ||
|
Methylation in Case |
9.27E-01 (Median) | Methylation in Control | 9.16E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon75 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg12993807) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:3.22E-02; Z-score:2.53E-01 | ||
|
Methylation in Case |
8.61E-01 (Median) | Methylation in Control | 8.32E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon76 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg13299707) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:3.39E-02; Z-score:2.65E-01 | ||
|
Methylation in Case |
8.00E-01 (Median) | Methylation in Control | 7.62E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon77 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg13301368) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.40E-02; Z-score:-2.31E-01 | ||
|
Methylation in Case |
9.54E-01 (Median) | Methylation in Control | 9.62E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon78 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg13376768) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.82E+00 | Statistic Test | p-value:3.58E-02; Z-score:-4.35E-01 | ||
|
Methylation in Case |
6.70E-02 (Median) | Methylation in Control | 1.22E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon79 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg13592947) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:3.72E-02; Z-score:7.07E-01 | ||
|
Methylation in Case |
8.67E-01 (Median) | Methylation in Control | 8.12E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon80 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg13681701) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:3.80E-02; Z-score:3.30E-01 | ||
|
Methylation in Case |
8.43E-01 (Median) | Methylation in Control | 8.17E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon81 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg14047153) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:4.24E-02; Z-score:1.81E-01 | ||
|
Methylation in Case |
9.06E-01 (Median) | Methylation in Control | 8.91E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon82 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg14596589) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:4.87E-02; Z-score:4.43E-01 | ||
|
Methylation in Case |
8.02E-01 (Median) | Methylation in Control | 7.54E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon83 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
Body (cg14671982) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.11E+00 | Statistic Test | p-value:4.94E-02; Z-score:4.99E-01 | ||
|
Methylation in Case |
1.06E-01 (Median) | Methylation in Control | 9.59E-02 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon84 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
3'UTR (cg10876737) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.46E+00 | Statistic Test | p-value:5.73E-12; Z-score:-2.05E+00 | ||
|
Methylation in Case |
4.48E-01 (Median) | Methylation in Control | 6.54E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon85 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
3'UTR (cg17568547) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:1.25E-10; Z-score:-2.39E+00 | ||
|
Methylation in Case |
8.44E-01 (Median) | Methylation in Control | 9.09E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon86 |
Methylation of SLC12A7 in atypical teratoid rhabdoid tumor | [ 13 ] | |||
|
Location |
3'UTR (cg19086001) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.31E+00 | Statistic Test | p-value:2.51E-10; Z-score:1.63E+00 | ||
|
Methylation in Case |
8.77E-01 (Median) | Methylation in Control | 6.70E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Depression |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A7 in depression | [ 14 ] | |||
|
Location |
Body (cg16017429) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:4.07E-03; Z-score:-4.05E-01 | ||
|
Methylation in Case |
9.37E-01 (Median) | Methylation in Control | 9.42E-01 (Median) | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A7 in depression | [ 14 ] | |||
|
Location |
Body (cg14671982) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:5.94E-03; Z-score:-6.06E-01 | ||
|
Methylation in Case |
7.69E-01 (Median) | Methylation in Control | 7.81E-01 (Median) | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A7 in depression | [ 14 ] | |||
|
Location |
Body (cg23998435) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:1.29E-02; Z-score:-3.53E-01 | ||
|
Methylation in Case |
9.18E-01 (Median) | Methylation in Control | 9.22E-01 (Median) | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A7 in depression | [ 14 ] | |||
|
Location |
Body (cg19854293) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.07E-02; Z-score:-5.03E-01 | ||
|
Methylation in Case |
7.80E-01 (Median) | Methylation in Control | 7.91E-01 (Median) | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A7 in depression | [ 14 ] | |||
|
Location |
Body (cg04213775) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.82E-02; Z-score:-5.17E-01 | ||
|
Methylation in Case |
7.79E-01 (Median) | Methylation in Control | 7.90E-01 (Median) | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A7 in depression | [ 14 ] | |||
|
Location |
Body (cg04380229) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:3.51E-02; Z-score:-3.12E-01 | ||
|
Methylation in Case |
7.72E-01 (Median) | Methylation in Control | 7.84E-01 (Median) | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A7 in depression | [ 14 ] | |||
|
Location |
Body (cg10489284) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:4.13E-02; Z-score:3.78E-01 | ||
|
Methylation in Case |
9.34E-01 (Median) | Methylation in Control | 9.28E-01 (Median) | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A7 in depression | [ 14 ] | |||
|
Location |
Body (cg00063291) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:4.25E-02; Z-score:-5.79E-01 | ||
|
Methylation in Case |
7.67E-01 (Median) | Methylation in Control | 7.80E-01 (Median) | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A7 in depression | [ 14 ] | |||
|
Location |
Body (cg03281154) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.47E-02; Z-score:-4.76E-01 | ||
|
Methylation in Case |
7.85E-01 (Median) | Methylation in Control | 7.95E-01 (Median) | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A7 in panic disorder | [ 15 ] | |||
|
Location |
Body (cg21946374) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-8.23E-01 | Statistic Test | p-value:3.27E-03; Z-score:-5.57E-01 | ||
|
Methylation in Case |
-1.30E+00 (Median) | Methylation in Control | -1.07E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A7 in panic disorder | [ 15 ] | |||
|
Location |
Body (cg22772691) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-8.77E-01 | Statistic Test | p-value:4.98E-03; Z-score:-3.67E-01 | ||
|
Methylation in Case |
-1.17E+00 (Median) | Methylation in Control | -1.03E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A7 in panic disorder | [ 15 ] | |||
|
Location |
Body (cg19773466) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-8.80E-01 | Statistic Test | p-value:6.07E-03; Z-score:-3.65E-01 | ||
|
Methylation in Case |
-6.61E-01 (Median) | Methylation in Control | -5.82E-01 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A7 in panic disorder | [ 15 ] | |||
|
Location |
Body (cg01266985) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:8.06E-03; Z-score:-5.19E-01 | ||
|
Methylation in Case |
2.24E+00 (Median) | Methylation in Control | 2.46E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A7 in panic disorder | [ 15 ] | |||
|
Location |
Body (cg18997983) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-8.73E-01 | Statistic Test | p-value:2.60E-02; Z-score:-2.71E-01 | ||
|
Methylation in Case |
-8.92E-01 (Median) | Methylation in Control | -7.79E-01 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A7 in panic disorder | [ 15 ] | |||
|
Location |
Body (cg23345930) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:2.78E-02; Z-score:3.66E-01 | ||
|
Methylation in Case |
5.51E+00 (Median) | Methylation in Control | 5.44E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A7 in panic disorder | [ 15 ] | |||
|
Location |
Body (cg02039689) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:2.99E-02; Z-score:1.65E-01 | ||
|
Methylation in Case |
3.61E+00 (Median) | Methylation in Control | 3.53E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lymphoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypermethylation of SLC12A7 in lymphoma than that in healthy individual | ||||
Studied Phenotype |
Lymphoma [ICD-11:2B30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:4.44E-13; Fold-change:0.412036394; Z-score:33.33417879 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
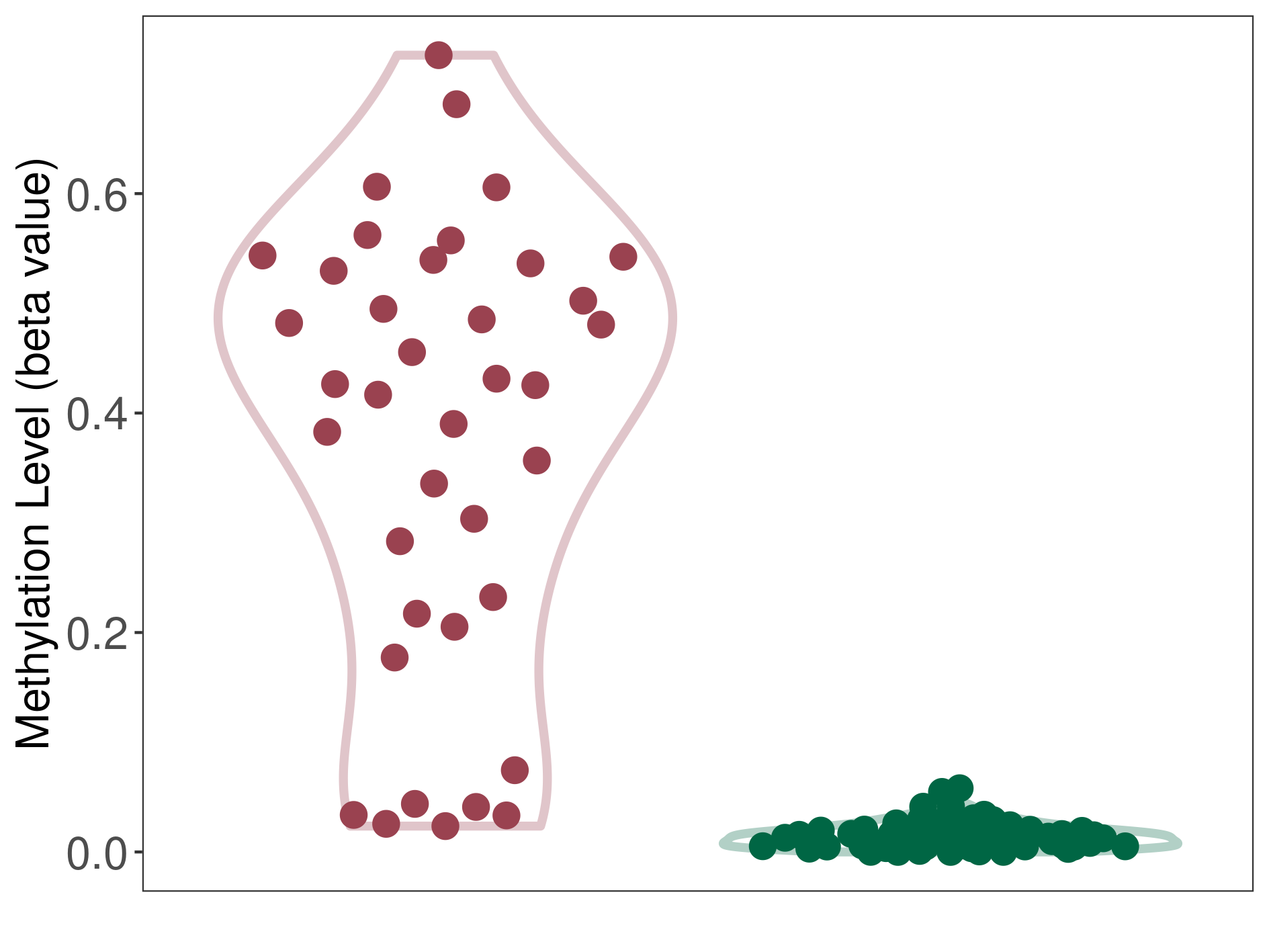
|
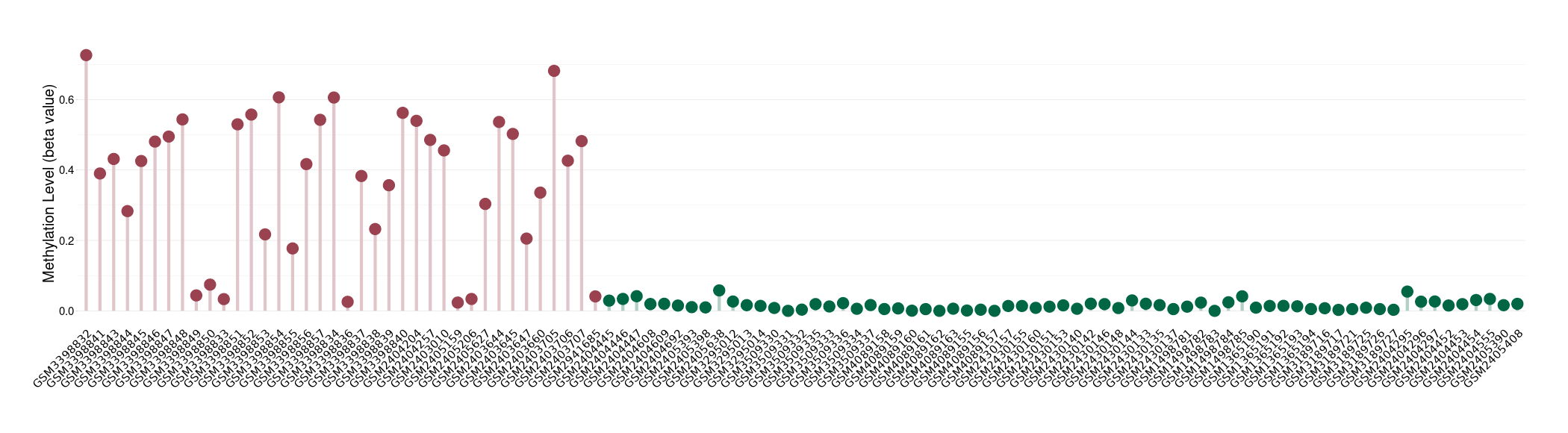 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
microRNA |
|||||
|
Unclear Phenotype |
101 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
let-7a directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
let-7a | miRNA Mature ID | let-7a-5p | ||
|
miRNA Sequence |
UGAGGUAGUAGGUUGUAUAGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon2 |
let-7b directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
let-7b | miRNA Mature ID | let-7b-5p | ||
|
miRNA Sequence |
UGAGGUAGUAGGUUGUGUGGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon3 |
let-7c directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
let-7c | miRNA Mature ID | let-7c-5p | ||
|
miRNA Sequence |
UGAGGUAGUAGGUUGUAUGGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon4 |
let-7d directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
let-7d | miRNA Mature ID | let-7d-5p | ||
|
miRNA Sequence |
AGAGGUAGUAGGUUGCAUAGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon5 |
let-7e directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
let-7e | miRNA Mature ID | let-7e-5p | ||
|
miRNA Sequence |
UGAGGUAGGAGGUUGUAUAGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon6 |
let-7f directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
let-7f | miRNA Mature ID | let-7f-5p | ||
|
miRNA Sequence |
UGAGGUAGUAGAUUGUAUAGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon7 |
let-7g directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
let-7g | miRNA Mature ID | let-7g-5p | ||
|
miRNA Sequence |
UGAGGUAGUAGUUUGUACAGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon8 |
let-7i directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
let-7i | miRNA Mature ID | let-7i-5p | ||
|
miRNA Sequence |
UGAGGUAGUAGUUUGUGCUGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon9 |
miR-1203 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1203 | miRNA Mature ID | miR-1203 | ||
|
miRNA Sequence |
CCCGGAGCCAGGAUGCAGCUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon10 |
miR-1291 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1291 | miRNA Mature ID | miR-1291 | ||
|
miRNA Sequence |
UGGCCCUGACUGAAGACCAGCAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon11 |
miR-130a directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-130a | miRNA Mature ID | miR-130a-3p | ||
|
miRNA Sequence |
CAGUGCAAUGUUAAAAGGGCAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon12 |
miR-130b directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-130b | miRNA Mature ID | miR-130b-3p | ||
|
miRNA Sequence |
CAGUGCAAUGAUGAAAGGGCAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon13 |
miR-146b directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-146b | miRNA Mature ID | miR-146b-3p | ||
|
miRNA Sequence |
GCCCUGUGGACUCAGUUCUGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon14 |
miR-148a directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-148a | miRNA Mature ID | miR-148a-3p | ||
|
miRNA Sequence |
UCAGUGCACUACAGAACUUUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon15 |
miR-148b directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-148b | miRNA Mature ID | miR-148b-3p | ||
|
miRNA Sequence |
UCAGUGCAUCACAGAACUUUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon16 |
miR-152 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-152 | miRNA Mature ID | miR-152-3p | ||
|
miRNA Sequence |
UCAGUGCAUGACAGAACUUGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon17 |
miR-1587 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1587 | miRNA Mature ID | miR-1587 | ||
|
miRNA Sequence |
UUGGGCUGGGCUGGGUUGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon18 |
miR-19a directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-19a | miRNA Mature ID | miR-19a-3p | ||
|
miRNA Sequence |
UGUGCAAAUCUAUGCAAAACUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon19 |
miR-19b directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-19b | miRNA Mature ID | miR-19b-3p | ||
|
miRNA Sequence |
UGUGCAAAUCCAUGCAAAACUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon20 |
miR-202 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-202 | miRNA Mature ID | miR-202-3p | ||
|
miRNA Sequence |
AGAGGUAUAGGGCAUGGGAA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon21 |
miR-223 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-223 | miRNA Mature ID | miR-223-3p | ||
|
miRNA Sequence |
UGUCAGUUUGUCAAAUACCCCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon22 |
miR-296 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-296 | miRNA Mature ID | miR-296-5p | ||
|
miRNA Sequence |
AGGGCCCCCCCUCAAUCCUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon23 |
miR-301a directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-301a | miRNA Mature ID | miR-301a-3p | ||
|
miRNA Sequence |
CAGUGCAAUAGUAUUGUCAAAGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon24 |
miR-301b directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-301b | miRNA Mature ID | miR-301b-3p | ||
|
miRNA Sequence |
CAGUGCAAUGAUAUUGUCAAAGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon25 |
miR-302c directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-302c | miRNA Mature ID | miR-302c-3p | ||
|
miRNA Sequence |
UAAGUGCUUCCAUGUUUCAGUGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon26 |
miR-3127 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3127 | miRNA Mature ID | miR-3127-3p | ||
|
miRNA Sequence |
UCCCCUUCUGCAGGCCUGCUGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon27 |
miR-3133 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3133 | miRNA Mature ID | miR-3133 | ||
|
miRNA Sequence |
UAAAGAACUCUUAAAACCCAAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon28 |
miR-3147 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3147 | miRNA Mature ID | miR-3147 | ||
|
miRNA Sequence |
GGUUGGGCAGUGAGGAGGGUGUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon29 |
miR-3180 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3180 | miRNA Mature ID | miR-3180-5p | ||
|
miRNA Sequence |
CUUCCAGACGCUCCGCCCCACGUCG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon30 |
miR-3194 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3194 | miRNA Mature ID | miR-3194-5p | ||
|
miRNA Sequence |
GGCCAGCCACCAGGAGGGCUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon31 |
miR-335 directly targets SLC12A7 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-335 | miRNA Mature ID | miR-335-5p | ||
|
miRNA Sequence |
UCAAGAGCAAUAACGAAAAAUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon32 |
miR-339 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-339 | miRNA Mature ID | miR-339-5p | ||
|
miRNA Sequence |
UCCCUGUCCUCCAGGAGCUCACG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon33 |
miR-33b directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-33b | miRNA Mature ID | miR-33b-3p | ||
|
miRNA Sequence |
CAGUGCCUCGGCAGUGCAGCCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon34 |
miR-3620 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3620 | miRNA Mature ID | miR-3620-5p | ||
|
miRNA Sequence |
GUGGGCUGGGCUGGGCUGGGCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon35 |
miR-3652 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3652 | miRNA Mature ID | miR-3652 | ||
|
miRNA Sequence |
CGGCUGGAGGUGUGAGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon36 |
miR-3666 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3666 | miRNA Mature ID | miR-3666 | ||
|
miRNA Sequence |
CAGUGCAAGUGUAGAUGCCGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon37 |
miR-4263 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4263 | miRNA Mature ID | miR-4263 | ||
|
miRNA Sequence |
AUUCUAAGUGCCUUGGCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon38 |
miR-4265 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4265 | miRNA Mature ID | miR-4265 | ||
|
miRNA Sequence |
CUGUGGGCUCAGCUCUGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon39 |
miR-4267 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4267 | miRNA Mature ID | miR-4267 | ||
|
miRNA Sequence |
UCCAGCUCGGUGGCAC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon40 |
miR-4276 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4276 | miRNA Mature ID | miR-4276 | ||
|
miRNA Sequence |
CUCAGUGACUCAUGUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon41 |
miR-4295 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4295 | miRNA Mature ID | miR-4295 | ||
|
miRNA Sequence |
CAGUGCAAUGUUUUCCUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon42 |
miR-4296 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4296 | miRNA Mature ID | miR-4296 | ||
|
miRNA Sequence |
AUGUGGGCUCAGGCUCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon43 |
miR-4312 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4312 | miRNA Mature ID | miR-4312 | ||
|
miRNA Sequence |
GGCCUUGUUCCUGUCCCCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon44 |
miR-4322 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4322 | miRNA Mature ID | miR-4322 | ||
|
miRNA Sequence |
CUGUGGGCUCAGCGCGUGGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon45 |
miR-4425 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4425 | miRNA Mature ID | miR-4425 | ||
|
miRNA Sequence |
UGUUGGGAUUCAGCAGGACCAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon46 |
miR-4430 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4430 | miRNA Mature ID | miR-4430 | ||
|
miRNA Sequence |
AGGCUGGAGUGAGCGGAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon47 |
miR-4458 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4458 | miRNA Mature ID | miR-4458 | ||
|
miRNA Sequence |
AGAGGUAGGUGUGGAAGAA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon48 |
miR-4492 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4492 | miRNA Mature ID | miR-4492 | ||
|
miRNA Sequence |
GGGGCUGGGCGCGCGCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon49 |
miR-4498 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4498 | miRNA Mature ID | miR-4498 | ||
|
miRNA Sequence |
UGGGCUGGCAGGGCAAGUGCUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon50 |
miR-4500 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4500 | miRNA Mature ID | miR-4500 | ||
|
miRNA Sequence |
UGAGGUAGUAGUUUCUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon51 |
miR-4505 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4505 | miRNA Mature ID | miR-4505 | ||
|
miRNA Sequence |
AGGCUGGGCUGGGACGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon52 |
miR-4509 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4509 | miRNA Mature ID | miR-4509 | ||
|
miRNA Sequence |
ACUAAAGGAUAUAGAAGGUUUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon53 |
miR-4511 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4511 | miRNA Mature ID | miR-4511 | ||
|
miRNA Sequence |
GAAGAACUGUUGCAUUUGCCCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon54 |
miR-454 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-454 | miRNA Mature ID | miR-454-3p | ||
|
miRNA Sequence |
UAGUGCAAUAUUGCUUAUAGGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon55 |
miR-4656 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4656 | miRNA Mature ID | miR-4656 | ||
|
miRNA Sequence |
UGGGCUGAGGGCAGGAGGCCUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon56 |
miR-4675 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4675 | miRNA Mature ID | miR-4675 | ||
|
miRNA Sequence |
GGGGCUGUGAUUGACCAGCAGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon57 |
miR-4691 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4691 | miRNA Mature ID | miR-4691-5p | ||
|
miRNA Sequence |
GUCCUCCAGGCCAUGAGCUGCGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon58 |
miR-4697 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4697 | miRNA Mature ID | miR-4697-3p | ||
|
miRNA Sequence |
UGUCAGUGACUCCUGCCCCUUGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon59 |
miR-4741 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4741 | miRNA Mature ID | miR-4741 | ||
|
miRNA Sequence |
CGGGCUGUCCGGAGGGGUCGGCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon60 |
miR-4744 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4744 | miRNA Mature ID | miR-4744 | ||
|
miRNA Sequence |
UCUAAAGACUAGACUUCGCUAUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon61 |
miR-4747 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4747 | miRNA Mature ID | miR-4747-3p | ||
|
miRNA Sequence |
AAGGCCCGGGCUUUCCUCCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon62 |
miR-4780 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4780 | miRNA Mature ID | miR-4780 | ||
|
miRNA Sequence |
ACCCUUGAGCCUGAUCCCUAGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon63 |
miR-4795 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4795 | miRNA Mature ID | miR-4795-5p | ||
|
miRNA Sequence |
AGAAGUGGCUAAUAAUAUUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon64 |
miR-4799 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4799 | miRNA Mature ID | miR-4799-5p | ||
|
miRNA Sequence |
AUCUAAAUGCAGCAUGCCAGUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon65 |
miR-488 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-488 | miRNA Mature ID | miR-488-3p | ||
|
miRNA Sequence |
UUGAAAGGCUAUUUCUUGGUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon66 |
miR-5001 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-5001 | miRNA Mature ID | miR-5001-5p | ||
|
miRNA Sequence |
AGGGCUGGACUCAGCGGCGGAGCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon67 |
miR-515 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-515 | miRNA Mature ID | miR-515-3p | ||
|
miRNA Sequence |
GAGUGCCUUCUUUUGGAGCGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon68 |
miR-518a directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-518a | miRNA Mature ID | miR-518a-5p | ||
|
miRNA Sequence |
CUGCAAAGGGAAGCCCUUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon69 |
miR-519a directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-519a | miRNA Mature ID | miR-519a-3p | ||
|
miRNA Sequence |
AAAGUGCAUCCUUUUAGAGUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon70 |
miR-519b directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-519b | miRNA Mature ID | miR-519b-3p | ||
|
miRNA Sequence |
AAAGUGCAUCCUUUUAGAGGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon71 |
miR-519c directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-519c | miRNA Mature ID | miR-519c-3p | ||
|
miRNA Sequence |
AAAGUGCAUCUUUUUAGAGGAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon72 |
miR-519e directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-519e | miRNA Mature ID | miR-519e-3p | ||
|
miRNA Sequence |
AAGUGCCUCCUUUUAGAGUGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon73 |
miR-520a directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-520a | miRNA Mature ID | miR-520a-5p | ||
|
miRNA Sequence |
CUCCAGAGGGAAGUACUUUCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon74 |
miR-520f directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-520f | miRNA Mature ID | miR-520f-3p | ||
|
miRNA Sequence |
AAGUGCUUCCUUUUAGAGGGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon75 |
miR-520g directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-520g | miRNA Mature ID | miR-520g-3p | ||
|
miRNA Sequence |
ACAAAGUGCUUCCCUUUAGAGUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon76 |
miR-520h directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-520h | miRNA Mature ID | miR-520h | ||
|
miRNA Sequence |
ACAAAGUGCUUCCCUUUAGAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon77 |
miR-525 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-525 | miRNA Mature ID | miR-525-5p | ||
|
miRNA Sequence |
CUCCAGAGGGAUGCACUUUCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon78 |
miR-527 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-527 | miRNA Mature ID | miR-527 | ||
|
miRNA Sequence |
CUGCAAAGGGAAGCCCUUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon79 |
miR-5586 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-5586 | miRNA Mature ID | miR-5586-5p | ||
|
miRNA Sequence |
UAUCCAGCUUGUUACUAUAUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon80 |
miR-576 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-576 | miRNA Mature ID | miR-576-5p | ||
|
miRNA Sequence |
AUUCUAAUUUCUCCACGUCUUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon81 |
miR-5787 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-5787 | miRNA Mature ID | miR-5787 | ||
|
miRNA Sequence |
GGGCUGGGGCGCGGGGAGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon82 |
miR-605 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-605 | miRNA Mature ID | miR-605-5p | ||
|
miRNA Sequence |
UAAAUCCCAUGGUGCCUUCUCCU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon83 |
miR-6124 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6124 | miRNA Mature ID | miR-6124 | ||
|
miRNA Sequence |
GGGAAAAGGAAGGGGGAGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon84 |
miR-651 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-651 | miRNA Mature ID | miR-651-3p | ||
|
miRNA Sequence |
AAAGGAAAGUGUAUCCUAAAAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon85 |
miR-6512 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6512 | miRNA Mature ID | miR-6512-3p | ||
|
miRNA Sequence |
UUCCAGCCCUUCUAAUGGUAGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon86 |
miR-6720 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6720 | miRNA Mature ID | miR-6720-5p | ||
|
miRNA Sequence |
UUCCAGCCCUGGUAGGCGCCGCG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon87 |
miR-6724 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6724 | miRNA Mature ID | miR-6724-5p | ||
|
miRNA Sequence |
CUGGGCCCGCGGCGGGCGUGGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon88 |
miR-6734 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6734 | miRNA Mature ID | miR-6734-3p | ||
|
miRNA Sequence |
CCCUUCCCUCACUCUUCUCUCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon89 |
miR-6756 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6756 | miRNA Mature ID | miR-6756-3p | ||
|
miRNA Sequence |
UCCCCUUCCUCCCUGCCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon90 |
miR-6773 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6773 | miRNA Mature ID | miR-6773-5p | ||
|
miRNA Sequence |
UUGGGCCCAGGAGUAAACAGGAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon91 |
miR-6775 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6775 | miRNA Mature ID | miR-6775-3p | ||
|
miRNA Sequence |
AGGCCCUGUCCUCUGCCCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon92 |
miR-6792 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6792 | miRNA Mature ID | miR-6792-3p | ||
|
miRNA Sequence |
CUCCUCCACAGCCCCUGCUCAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon93 |
miR-6829 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6829 | miRNA Mature ID | miR-6829-5p | ||
|
miRNA Sequence |
UGGGCUGCUGAGAAGGGGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon94 |
miR-6830 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6830 | miRNA Mature ID | miR-6830-5p | ||
|
miRNA Sequence |
CCAAGGAAGGAGGCUGGACAUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon95 |
miR-6842 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6842 | miRNA Mature ID | miR-6842-3p | ||
|
miRNA Sequence |
UUGGCUGGUCUCUGCUCCGCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon96 |
miR-6849 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6849 | miRNA Mature ID | miR-6849-3p | ||
|
miRNA Sequence |
ACCAGCCUGUGUCCACCUCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon97 |
miR-6851 directly targets SLC12A7 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6851 | miRNA Mature ID | miR-6851-3p | ||
|
miRNA Sequence |
UGGCCCUUUGUACCCCUCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon98 |
miR-762 directly targets SLC12A7 | [ 17 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-762 | miRNA Mature ID | miR-762 | ||
|
miRNA Sequence |
GGGGCUGGGGCCGGGGCCGAGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon99 |
miR-766 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-766 | miRNA Mature ID | miR-766-3p | ||
|
miRNA Sequence |
ACUCCAGCCCCACAGCCUCAGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon100 |
miR-7976 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-7976 | miRNA Mature ID | miR-7976 | ||
|
miRNA Sequence |
UGCCCUGAGACUUUUGCUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon101 |
miR-98 directly targets SLC12A7 | [ 16 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-98 | miRNA Mature ID | miR-98-5p | ||
|
miRNA Sequence |
UGAGGUAGUAAGUUGUAUUGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
