Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0086 Transporter Info | ||||
| Gene Name | SLC12A5 | ||||
| Transporter Name | Electroneutral potassium-chloride cotransporter 2 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Colon cancer |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg17846334) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:3.05E+00 | Statistic Test | p-value:2.13E-08; Z-score:6.03E+00 | ||
|
Methylation in Case |
4.75E-01 (Median) | Methylation in Control | 1.56E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
5'UTR (cg22115748) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:2.90E-03; Z-score:-1.99E+00 | ||
|
Methylation in Case |
4.70E-01 (Median) | Methylation in Control | 5.45E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg06750832) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.57E+00 | Statistic Test | p-value:1.06E-08; Z-score:2.40E+00 | ||
|
Methylation in Case |
5.38E-01 (Median) | Methylation in Control | 2.09E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
TSS200 (cg22988581) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:2.96E-03; Z-score:-1.32E+00 | ||
|
Methylation in Case |
2.90E-01 (Median) | Methylation in Control | 3.43E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
1stExon (cg16435571) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.58E+00 | Statistic Test | p-value:5.37E-07; Z-score:2.31E+00 | ||
|
Methylation in Case |
4.99E-01 (Median) | Methylation in Control | 3.17E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A5 in colon adenocarcinoma | [ 1 ] | |||
|
Location |
1stExon (cg22049569) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.87E+00 | Statistic Test | p-value:2.52E-06; Z-score:2.34E+00 | ||
|
Methylation in Case |
4.00E-01 (Median) | Methylation in Control | 2.13E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
9 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
5'UTR (cg06852309) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:2.60E-13; Z-score:-2.72E+00 | ||
|
Methylation in Case |
4.36E-01 (Median) | Methylation in Control | 5.93E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS1500 (cg09595245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:1.39E-04; Z-score:-7.08E-01 | ||
|
Methylation in Case |
7.16E-01 (Median) | Methylation in Control | 7.86E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg21609339) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.61E+00 | Statistic Test | p-value:2.72E-06; Z-score:4.82E+00 | ||
|
Methylation in Case |
3.39E-01 (Median) | Methylation in Control | 2.11E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg23463144) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.32E+00 | Statistic Test | p-value:2.25E-05; Z-score:1.49E+00 | ||
|
Methylation in Case |
9.72E-02 (Median) | Methylation in Control | 7.35E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
TSS200 (cg04980291) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:1.15E-04; Z-score:5.87E-01 | ||
|
Methylation in Case |
1.09E-01 (Median) | Methylation in Control | 9.59E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
1stExon (cg14921743) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.38E+00 | Statistic Test | p-value:1.54E-06; Z-score:1.69E+00 | ||
|
Methylation in Case |
5.92E-02 (Median) | Methylation in Control | 2.49E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg05654361) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.29E+00 | Statistic Test | p-value:4.47E-10; Z-score:-4.78E+00 | ||
|
Methylation in Case |
5.60E-01 (Median) | Methylation in Control | 7.21E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg01160557) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.98E+00 | Statistic Test | p-value:1.62E-09; Z-score:-2.01E+00 | ||
|
Methylation in Case |
1.55E-01 (Median) | Methylation in Control | 3.07E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A5 in hepatocellular carcinoma | [ 2 ] | |||
|
Location |
Body (cg12832857) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:2.51E-02; Z-score:-7.59E-02 | ||
|
Methylation in Case |
7.97E-02 (Median) | Methylation in Control | 8.22E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Pancretic ductal adenocarcinoma |
8 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
5'UTR (cg19755318) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.49E+00 | Statistic Test | p-value:2.93E-16; Z-score:1.77E+00 | ||
|
Methylation in Case |
1.77E-01 (Median) | Methylation in Control | 1.19E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
5'UTR (cg19464780) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:9.82E-09; Z-score:-1.58E+00 | ||
|
Methylation in Case |
8.01E-01 (Median) | Methylation in Control | 8.61E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
TSS200 (cg08288703) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:1.18E-09; Z-score:-1.45E+00 | ||
|
Methylation in Case |
3.98E-01 (Median) | Methylation in Control | 4.85E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
TSS200 (cg24707219) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:5.54E-03; Z-score:-3.67E-01 | ||
|
Methylation in Case |
5.71E-01 (Median) | Methylation in Control | 6.01E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
Body (cg25945642) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:1.81E-09; Z-score:-1.81E+00 | ||
|
Methylation in Case |
5.84E-01 (Median) | Methylation in Control | 7.11E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
Body (cg17332016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:1.29E-04; Z-score:-2.90E-01 | ||
|
Methylation in Case |
5.31E-02 (Median) | Methylation in Control | 5.73E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
Body (cg04135385) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:1.94E-02; Z-score:8.03E-01 | ||
|
Methylation in Case |
8.50E-01 (Median) | Methylation in Control | 8.20E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A5 in pancretic ductal adenocarcinoma | [ 3 ] | |||
|
Location |
Body (cg11244695) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:4.88E-02; Z-score:-3.75E-01 | ||
|
Methylation in Case |
8.84E-01 (Median) | Methylation in Control | 8.99E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A5 in prostate cancer | [ 4 ] | |||
|
Location |
5'UTR (cg03363743) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.71E+00 | Statistic Test | p-value:5.95E-03; Z-score:2.65E+00 | ||
|
Methylation in Case |
5.81E-01 (Median) | Methylation in Control | 3.41E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A5 in prostate cancer | [ 4 ] | |||
|
Location |
TSS200 (cg25963505) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:5.80E-03; Z-score:2.71E+00 | ||
|
Methylation in Case |
8.80E-01 (Median) | Methylation in Control | 7.34E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A5 in prostate cancer | [ 4 ] | |||
|
Location |
1stExon (cg13348944) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.95E+00 | Statistic Test | p-value:1.16E-02; Z-score:7.27E+00 | ||
|
Methylation in Case |
6.24E-01 (Median) | Methylation in Control | 3.20E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A5 in prostate cancer | [ 4 ] | |||
|
Location |
Body (cg22882121) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.50E+00 | Statistic Test | p-value:1.32E-02; Z-score:-2.54E+00 | ||
|
Methylation in Case |
4.36E-01 (Median) | Methylation in Control | 6.52E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
10 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A5 in bladder cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg04539957) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.28E+00 | Statistic Test | p-value:1.67E-08; Z-score:6.73E+00 | ||
|
Methylation in Case |
7.48E-01 (Median) | Methylation in Control | 5.86E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A5 in bladder cancer | [ 5 ] | |||
|
Location |
TSS1500 (cg09595245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.20E+00 | Statistic Test | p-value:7.80E-05; Z-score:-4.94E+00 | ||
|
Methylation in Case |
7.16E-01 (Median) | Methylation in Control | 8.61E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A5 in bladder cancer | [ 5 ] | |||
|
Location |
TSS200 (cg17920479) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.21E+00 | Statistic Test | p-value:3.94E-09; Z-score:9.95E+00 | ||
|
Methylation in Case |
4.54E-01 (Median) | Methylation in Control | 2.05E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A5 in bladder cancer | [ 5 ] | |||
|
Location |
TSS200 (cg07191594) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.81E+00 | Statistic Test | p-value:2.62E-07; Z-score:1.06E+01 | ||
|
Methylation in Case |
3.99E-01 (Median) | Methylation in Control | 2.20E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A5 in bladder cancer | [ 5 ] | |||
|
Location |
TSS200 (cg21609339) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.85E+00 | Statistic Test | p-value:9.28E-06; Z-score:7.35E+00 | ||
|
Methylation in Case |
3.16E-01 (Median) | Methylation in Control | 1.71E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A5 in bladder cancer | [ 5 ] | |||
|
Location |
TSS200 (cg23463144) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.58E+00 | Statistic Test | p-value:9.05E-05; Z-score:9.25E+00 | ||
|
Methylation in Case |
6.82E-02 (Median) | Methylation in Control | 4.32E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A5 in bladder cancer | [ 5 ] | |||
|
Location |
TSS200 (cg04980291) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.34E+00 | Statistic Test | p-value:5.76E-04; Z-score:2.73E+00 | ||
|
Methylation in Case |
9.84E-02 (Median) | Methylation in Control | 7.37E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A5 in bladder cancer | [ 5 ] | |||
|
Location |
1stExon (cg14921743) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.73E+00 | Statistic Test | p-value:2.45E-04; Z-score:8.17E+00 | ||
|
Methylation in Case |
7.53E-02 (Median) | Methylation in Control | 2.76E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A5 in bladder cancer | [ 5 ] | |||
|
Location |
Body (cg23274660) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-4.26E+00 | Statistic Test | p-value:3.22E-08; Z-score:-8.72E+00 | ||
|
Methylation in Case |
9.33E-02 (Median) | Methylation in Control | 3.98E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC12A5 in bladder cancer | [ 5 ] | |||
|
Location |
Body (cg01160557) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:2.36E-03; Z-score:-2.15E+00 | ||
|
Methylation in Case |
8.41E-02 (Median) | Methylation in Control | 1.15E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
11 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A5 in breast cancer | [ 6 ] | |||
|
Location |
TSS1500 (cg04539957) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:6.56E-13; Z-score:1.76E+00 | ||
|
Methylation in Case |
7.18E-01 (Median) | Methylation in Control | 6.23E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A5 in breast cancer | [ 6 ] | |||
|
Location |
TSS200 (cg17920479) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.75E+00 | Statistic Test | p-value:1.16E-17; Z-score:4.10E+00 | ||
|
Methylation in Case |
3.98E-01 (Median) | Methylation in Control | 2.28E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A5 in breast cancer | [ 6 ] | |||
|
Location |
TSS200 (cg07191594) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.58E+00 | Statistic Test | p-value:1.93E-17; Z-score:3.37E+00 | ||
|
Methylation in Case |
3.59E-01 (Median) | Methylation in Control | 2.27E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A5 in breast cancer | [ 6 ] | |||
|
Location |
TSS200 (cg21609339) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.55E+00 | Statistic Test | p-value:2.02E-13; Z-score:3.12E+00 | ||
|
Methylation in Case |
2.71E-01 (Median) | Methylation in Control | 1.75E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A5 in breast cancer | [ 6 ] | |||
|
Location |
TSS200 (cg04980291) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.26E+00 | Statistic Test | p-value:8.77E-06; Z-score:7.89E-01 | ||
|
Methylation in Case |
9.86E-02 (Median) | Methylation in Control | 7.84E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A5 in breast cancer | [ 6 ] | |||
|
Location |
TSS200 (cg23463144) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.28E+00 | Statistic Test | p-value:9.81E-06; Z-score:9.26E-01 | ||
|
Methylation in Case |
6.60E-02 (Median) | Methylation in Control | 5.16E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A5 in breast cancer | [ 6 ] | |||
|
Location |
1stExon (cg14921743) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.38E+00 | Statistic Test | p-value:4.25E-07; Z-score:2.03E-01 | ||
|
Methylation in Case |
2.56E-02 (Median) | Methylation in Control | 1.86E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A5 in breast cancer | [ 6 ] | |||
|
Location |
Body (cg23274660) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:1.60E-05; Z-score:-8.88E-01 | ||
|
Methylation in Case |
1.83E-01 (Median) | Methylation in Control | 2.47E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A5 in breast cancer | [ 6 ] | |||
|
Location |
Body (cg01160557) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.75E+00 | Statistic Test | p-value:1.61E-03; Z-score:-1.46E+00 | ||
|
Methylation in Case |
9.54E-02 (Median) | Methylation in Control | 1.67E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC12A5 in breast cancer | [ 6 ] | |||
|
Location |
Body (cg23508607) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.85E-02; Z-score:-3.48E-02 | ||
|
Methylation in Case |
6.96E-02 (Median) | Methylation in Control | 7.02E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC12A5 in breast cancer | [ 6 ] | |||
|
Location |
Body (cg12832857) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.32E+00 | Statistic Test | p-value:4.18E-02; Z-score:4.60E-01 | ||
|
Methylation in Case |
7.68E-02 (Median) | Methylation in Control | 5.82E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A5 in clear cell renal cell carcinoma | [ 7 ] | |||
|
Location |
TSS1500 (cg04539957) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:1.45E-07; Z-score:2.89E+00 | ||
|
Methylation in Case |
8.51E-01 (Median) | Methylation in Control | 7.26E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A5 in clear cell renal cell carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg07191594) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.88E+00 | Statistic Test | p-value:3.79E-12; Z-score:4.28E+00 | ||
|
Methylation in Case |
3.65E-01 (Median) | Methylation in Control | 1.94E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A5 in clear cell renal cell carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg17920479) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.16E+00 | Statistic Test | p-value:1.50E-10; Z-score:4.51E+00 | ||
|
Methylation in Case |
4.08E-01 (Median) | Methylation in Control | 1.89E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A5 in clear cell renal cell carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg21609339) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.40E+00 | Statistic Test | p-value:2.53E-06; Z-score:1.75E+00 | ||
|
Methylation in Case |
2.14E-01 (Median) | Methylation in Control | 1.53E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A5 in clear cell renal cell carcinoma | [ 7 ] | |||
|
Location |
TSS200 (cg23463144) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.30E+00 | Statistic Test | p-value:1.97E-02; Z-score:9.51E-01 | ||
|
Methylation in Case |
2.52E-02 (Median) | Methylation in Control | 1.94E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A5 in clear cell renal cell carcinoma | [ 7 ] | |||
|
Location |
1stExon (cg14921743) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.00E+00 | Statistic Test | p-value:2.02E-02; Z-score:4.42E-03 | ||
|
Methylation in Case |
1.71E-02 (Median) | Methylation in Control | 1.71E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
12 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A5 in colorectal cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg09595245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.12E+00 | Statistic Test | p-value:3.69E-07; Z-score:-2.29E+00 | ||
|
Methylation in Case |
8.06E-01 (Median) | Methylation in Control | 9.03E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A5 in colorectal cancer | [ 8 ] | |||
|
Location |
TSS200 (cg21609339) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.92E+00 | Statistic Test | p-value:1.71E-13; Z-score:4.62E+00 | ||
|
Methylation in Case |
5.79E-01 (Median) | Methylation in Control | 3.02E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A5 in colorectal cancer | [ 8 ] | |||
|
Location |
TSS200 (cg17920479) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.40E+00 | Statistic Test | p-value:1.67E-08; Z-score:2.06E+00 | ||
|
Methylation in Case |
6.57E-01 (Median) | Methylation in Control | 4.69E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A5 in colorectal cancer | [ 8 ] | |||
|
Location |
TSS200 (cg04980291) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.15E+00 | Statistic Test | p-value:6.40E-08; Z-score:3.83E-01 | ||
|
Methylation in Case |
1.80E-01 (Median) | Methylation in Control | 1.57E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A5 in colorectal cancer | [ 8 ] | |||
|
Location |
TSS200 (cg23463144) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:3.24E-06; Z-score:1.02E+00 | ||
|
Methylation in Case |
1.53E-01 (Median) | Methylation in Control | 1.26E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A5 in colorectal cancer | [ 8 ] | |||
|
Location |
TSS200 (cg07191594) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:3.96E-03; Z-score:7.12E-01 | ||
|
Methylation in Case |
6.28E-01 (Median) | Methylation in Control | 5.55E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A5 in colorectal cancer | [ 8 ] | |||
|
Location |
1stExon (cg14921743) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.10E+01 | Statistic Test | p-value:3.26E-13; Z-score:2.56E+01 | ||
|
Methylation in Case |
4.18E-01 (Median) | Methylation in Control | 1.99E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC12A5 in colorectal cancer | [ 8 ] | |||
|
Location |
Body (cg01634998) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:9.11E-04; Z-score:1.34E-01 | ||
|
Methylation in Case |
2.12E-01 (Median) | Methylation in Control | 2.10E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC12A5 in colorectal cancer | [ 8 ] | |||
|
Location |
Body (cg23508607) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.18E+00 | Statistic Test | p-value:7.36E-03; Z-score:-1.08E+00 | ||
|
Methylation in Case |
1.22E-01 (Median) | Methylation in Control | 1.44E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC12A5 in colorectal cancer | [ 8 ] | |||
|
Location |
Body (cg12832857) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:1.97E-02; Z-score:5.57E-01 | ||
|
Methylation in Case |
5.37E-02 (Median) | Methylation in Control | 4.49E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC12A5 in colorectal cancer | [ 8 ] | |||
|
Location |
Body (cg23274660) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.25E+00 | Statistic Test | p-value:2.31E-02; Z-score:-6.38E-01 | ||
|
Methylation in Case |
2.65E-01 (Median) | Methylation in Control | 3.32E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
TSS1500 (cg09595245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:4.35E-02; Z-score:7.67E-01 | ||
|
Methylation in Case |
8.27E-01 (Median) | Methylation in Control | 8.06E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg01160557) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:1.02E-03; Z-score:-1.46E+00 | ||
|
Methylation in Case |
1.22E-01 (Median) | Methylation in Control | 1.65E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A5 in lung adenocarcinoma | [ 9 ] | |||
|
Location |
Body (cg23274660) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:1.83E-02; Z-score:-1.44E+00 | ||
|
Methylation in Case |
3.23E-01 (Median) | Methylation in Control | 4.36E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A5 in papillary thyroid cancer | [ 10 ] | |||
|
Location |
TSS1500 (cg04539957) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:7.72E-03; Z-score:8.89E-01 | ||
|
Methylation in Case |
6.02E-01 (Median) | Methylation in Control | 5.60E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A5 in papillary thyroid cancer | [ 10 ] | |||
|
Location |
TSS200 (cg07191594) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:1.82E-05; Z-score:8.54E-01 | ||
|
Methylation in Case |
1.78E-01 (Median) | Methylation in Control | 1.52E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A5 in papillary thyroid cancer | [ 10 ] | |||
|
Location |
TSS200 (cg17920479) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.23E+00 | Statistic Test | p-value:1.43E-04; Z-score:9.97E-01 | ||
|
Methylation in Case |
1.79E-01 (Median) | Methylation in Control | 1.46E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A5 in papillary thyroid cancer | [ 10 ] | |||
|
Location |
TSS200 (cg21609339) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:6.44E-04; Z-score:9.41E-01 | ||
|
Methylation in Case |
1.22E-01 (Median) | Methylation in Control | 1.04E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A5 in papillary thyroid cancer | [ 10 ] | |||
|
Location |
TSS200 (cg23463144) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:2.15E-02; Z-score:2.87E-01 | ||
|
Methylation in Case |
5.77E-02 (Median) | Methylation in Control | 5.44E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A5 in papillary thyroid cancer | [ 10 ] | |||
|
Location |
Body (cg23274660) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.28E+00 | Statistic Test | p-value:1.21E-06; Z-score:-1.32E+00 | ||
|
Methylation in Case |
3.94E-01 (Median) | Methylation in Control | 5.05E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC12A5 in papillary thyroid cancer | [ 10 ] | |||
|
Location |
Body (cg01160557) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:3.13E-02; Z-score:-1.19E+00 | ||
|
Methylation in Case |
1.22E-01 (Median) | Methylation in Control | 1.66E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Systemic lupus erythematosus |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A5 in systemic lupus erythematosus | [ 11 ] | |||
|
Location |
TSS1500 (cg09595245) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:7.26E-04; Z-score:-6.48E-02 | ||
|
Methylation in Case |
8.98E-01 (Median) | Methylation in Control | 9.01E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A5 in systemic lupus erythematosus | [ 11 ] | |||
|
Location |
TSS200 (cg04980291) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:1.23E-02; Z-score:-1.18E-01 | ||
|
Methylation in Case |
1.06E-01 (Median) | Methylation in Control | 1.11E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A5 in systemic lupus erythematosus | [ 11 ] | |||
|
Location |
Body (cg23274660) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:7.75E-04; Z-score:-3.28E-01 | ||
|
Methylation in Case |
3.98E-01 (Median) | Methylation in Control | 4.36E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A5 in systemic lupus erythematosus | [ 11 ] | |||
|
Location |
Body (cg23508607) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:8.44E-03; Z-score:-2.72E-01 | ||
|
Methylation in Case |
8.16E-02 (Median) | Methylation in Control | 8.73E-02 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Depression |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A5 in depression | [ 12 ] | |||
|
Location |
TSS200 (cg17920479) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:1.02E-02; Z-score:6.60E-01 | ||
|
Methylation in Case |
1.38E-01 (Median) | Methylation in Control | 1.27E-01 (Median) | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A5 in depression | [ 12 ] | |||
|
Location |
Body (cg23274660) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.73E-02; Z-score:-3.87E-01 | ||
|
Methylation in Case |
3.37E-01 (Median) | Methylation in Control | 3.54E-01 (Median) | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
HIV infection |
6 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A5 in HIV infection | [ 13 ] | |||
|
Location |
TSS200 (cg17920479) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.30E+00 | Statistic Test | p-value:1.24E-05; Z-score:1.63E+00 | ||
|
Methylation in Case |
2.43E-01 (Median) | Methylation in Control | 1.86E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A5 in HIV infection | [ 13 ] | |||
|
Location |
TSS200 (cg07191594) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.30E+00 | Statistic Test | p-value:1.45E-05; Z-score:1.69E+00 | ||
|
Methylation in Case |
2.74E-01 (Median) | Methylation in Control | 2.11E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A5 in HIV infection | [ 13 ] | |||
|
Location |
TSS200 (cg21609339) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.50E+00 | Statistic Test | p-value:1.48E-05; Z-score:2.40E+00 | ||
|
Methylation in Case |
2.07E-01 (Median) | Methylation in Control | 1.38E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A5 in HIV infection | [ 13 ] | |||
|
Location |
1stExon (cg14921743) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.30E+00 | Statistic Test | p-value:1.67E-02; Z-score:6.53E-01 | ||
|
Methylation in Case |
1.99E-02 (Median) | Methylation in Control | 1.53E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A5 in HIV infection | [ 13 ] | |||
|
Location |
Body (cg01160557) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:3.38E-04; Z-score:-4.93E-01 | ||
|
Methylation in Case |
9.34E-02 (Median) | Methylation in Control | 1.04E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC12A5 in HIV infection | [ 13 ] | |||
|
Location |
Body (cg23274660) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:1.90E-03; Z-score:-7.73E-01 | ||
|
Methylation in Case |
3.53E-01 (Median) | Methylation in Control | 4.04E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A5 in panic disorder | [ 14 ] | |||
|
Location |
TSS200 (cg07191594) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-8.92E-01 | Statistic Test | p-value:1.45E-03; Z-score:-6.20E-01 | ||
|
Methylation in Case |
-2.75E+00 (Median) | Methylation in Control | -2.46E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A5 in panic disorder | [ 14 ] | |||
|
Location |
TSS200 (cg17920479) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-9.37E-01 | Statistic Test | p-value:1.74E-02; Z-score:-3.60E-01 | ||
|
Methylation in Case |
-2.94E+00 (Median) | Methylation in Control | -2.75E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A5 in panic disorder | [ 14 ] | |||
|
Location |
TSS200 (cg21609339) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-9.61E-01 | Statistic Test | p-value:2.42E-02; Z-score:-2.70E-01 | ||
|
Methylation in Case |
-3.22E+00 (Median) | Methylation in Control | -3.10E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A5 in panic disorder | [ 14 ] | |||
|
Location |
Body (cg01160557) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:9.51E-01 | Statistic Test | p-value:2.80E-04; Z-score:5.05E-01 | ||
|
Methylation in Case |
-3.84E+00 (Median) | Methylation in Control | -4.04E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC12A5 in panic disorder | [ 14 ] | |||
|
Location |
Body (cg23274660) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:7.76E-01 | Statistic Test | p-value:3.01E-03; Z-score:5.65E-01 | ||
|
Methylation in Case |
-8.55E-01 (Median) | Methylation in Control | -1.10E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Atypical teratoid rhabdoid tumor |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC12A5 in atypical teratoid rhabdoid tumor | [ 15 ] | |||
|
Location |
1stExon (cg14921743) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.26E+00 | Statistic Test | p-value:7.07E-19; Z-score:3.54E+00 | ||
|
Methylation in Case |
5.61E-01 (Median) | Methylation in Control | 2.48E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC12A5 in atypical teratoid rhabdoid tumor | [ 15 ] | |||
|
Location |
Body (cg01160557) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:4.91E-05; Z-score:-9.85E-01 | ||
|
Methylation in Case |
8.17E-01 (Median) | Methylation in Control | 8.98E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC12A5 in atypical teratoid rhabdoid tumor | [ 15 ] | |||
|
Location |
Body (cg01634998) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.99E+00 | Statistic Test | p-value:7.46E-05; Z-score:-1.24E+00 | ||
|
Methylation in Case |
1.05E-01 (Median) | Methylation in Control | 2.09E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC12A5 in atypical teratoid rhabdoid tumor | [ 15 ] | |||
|
Location |
Body (cg12832857) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:2.98E-02; Z-score:6.35E-02 | ||
|
Methylation in Case |
7.71E-02 (Median) | Methylation in Control | 7.65E-02 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Squamous cell carcinoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of SLC12A5 in squamous cell carcinoma than that in healthy individual | ||||
Studied Phenotype |
Squamous cell carcinoma [ICD-11:2B60] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.005539222; Fold-change:-0.259128293; Z-score:-1.481890268 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue adjacent to the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
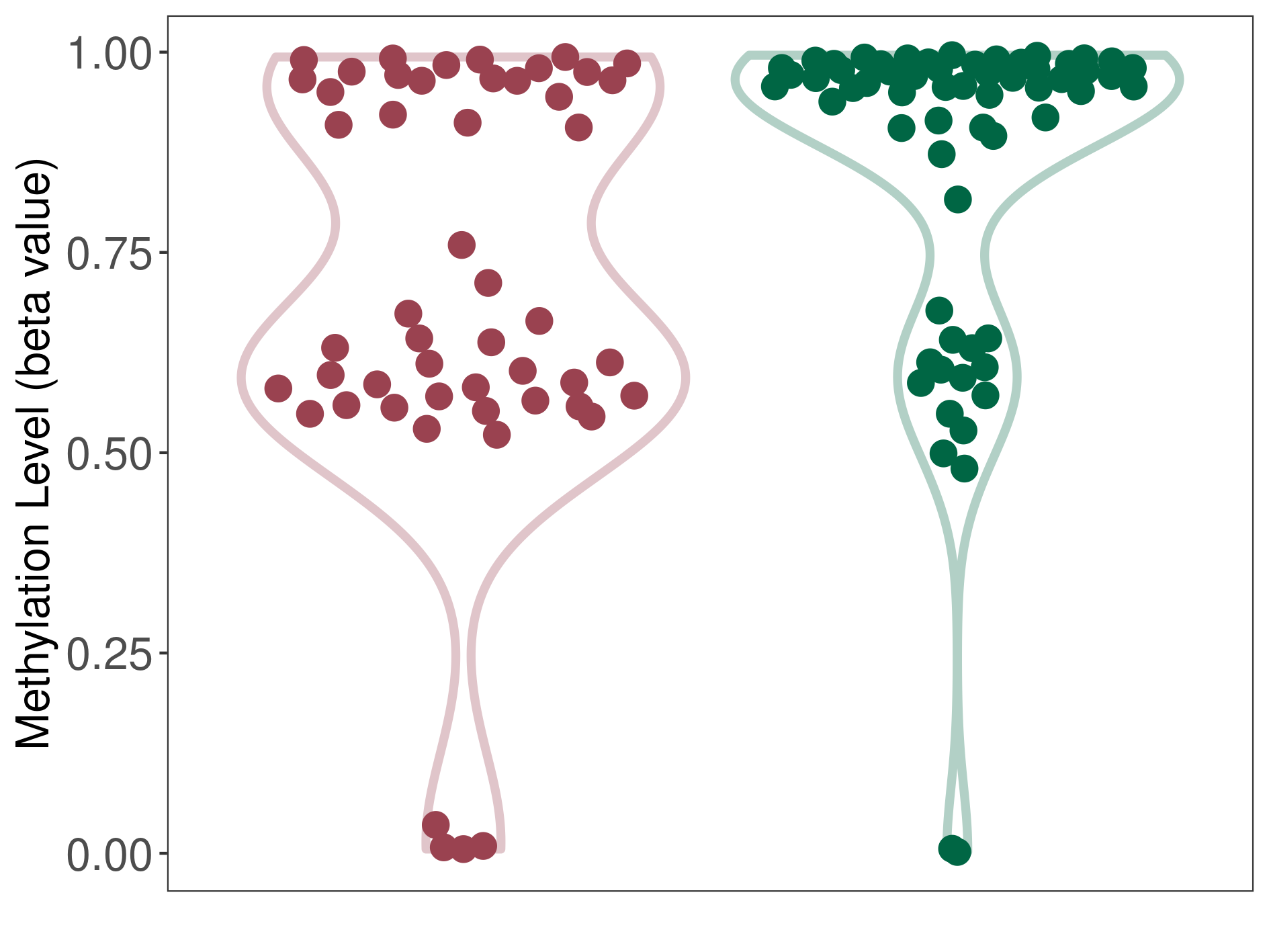
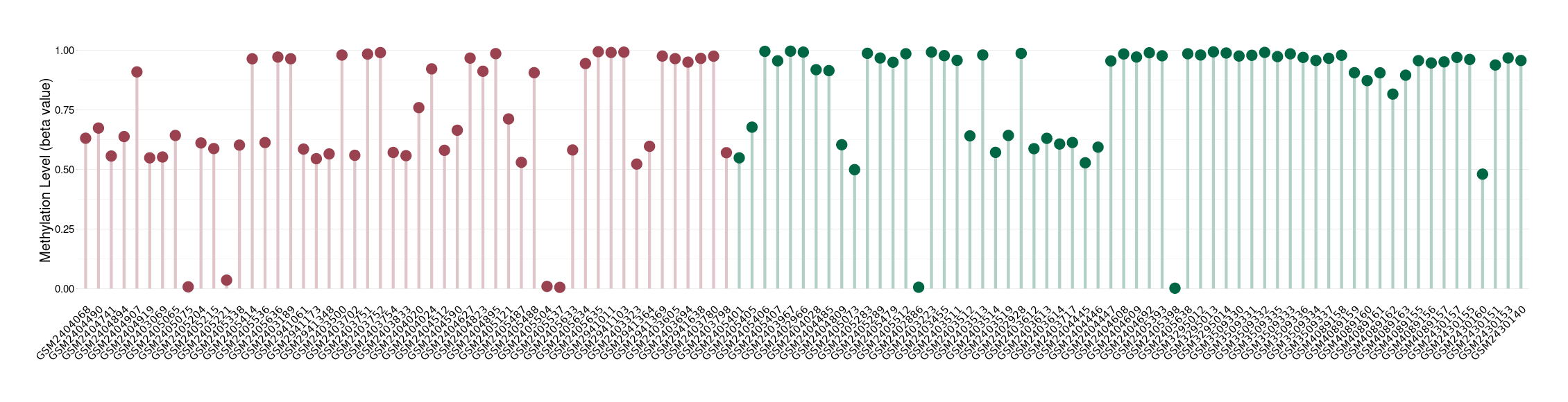
Brain neuroepithelial tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC12A5 in brain neuroepithelial tumour than that in healthy individual | ||||
Studied Phenotype |
Brain neuroepithelial tumour [ICD-11:2A00.2Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.003272911; Fold-change:-0.314442811; Z-score:-1.40310293 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
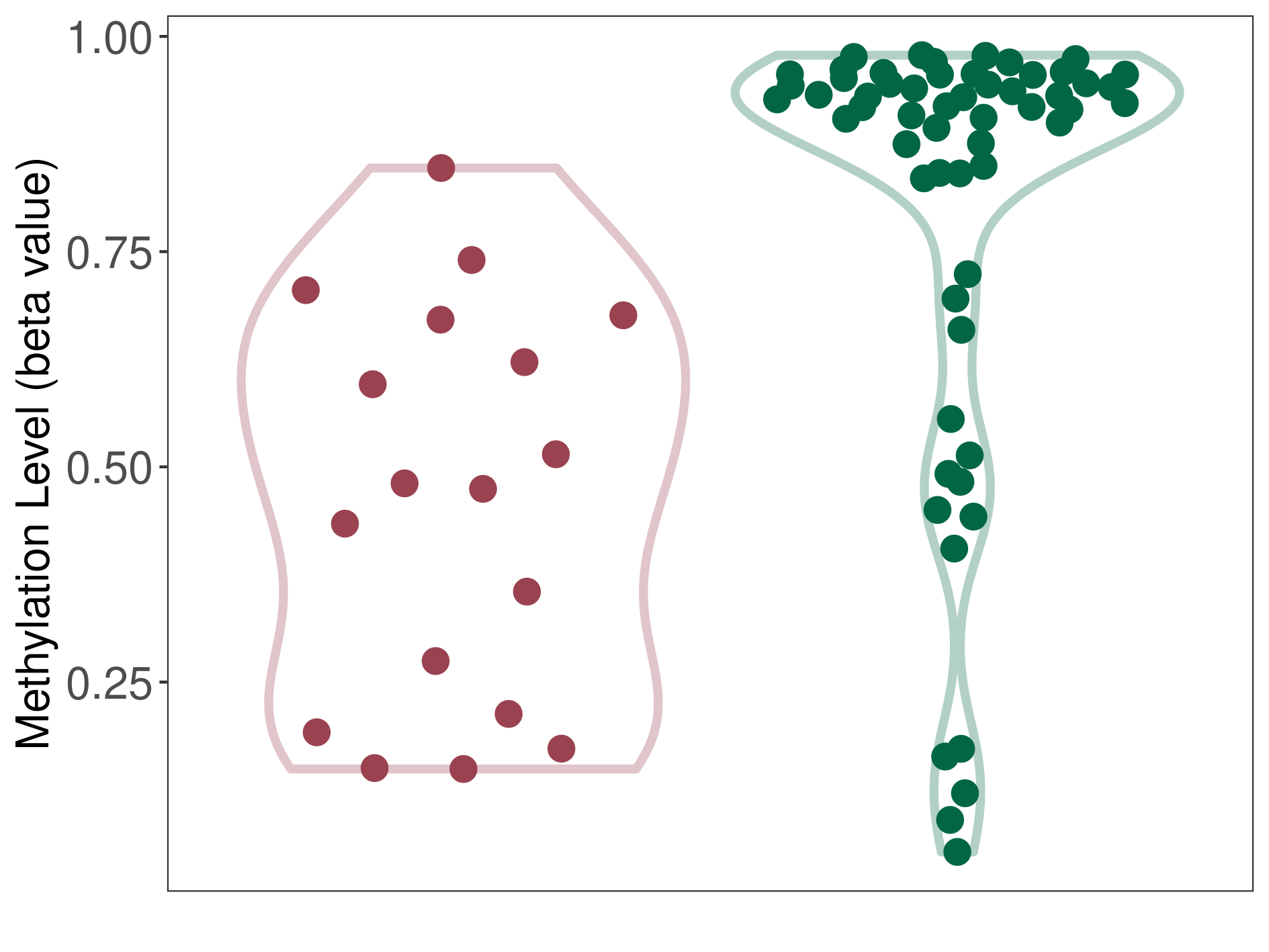
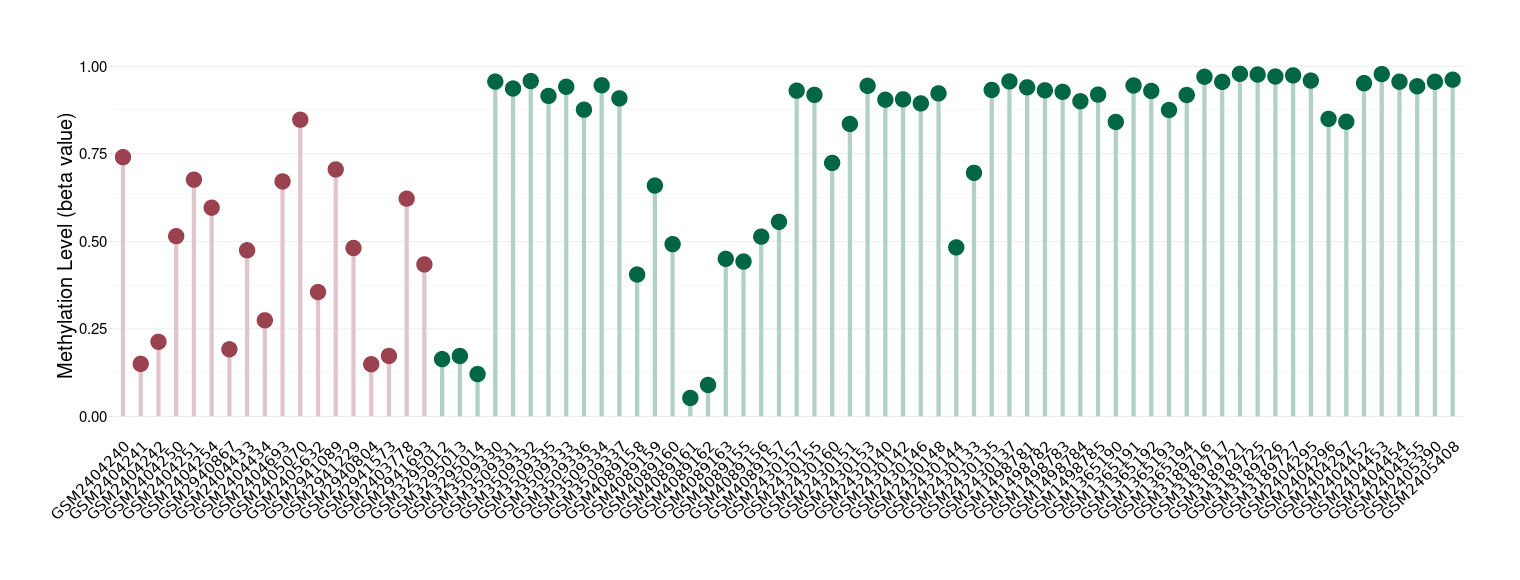
Melanoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC12A5 in melanoma than that in healthy individual | ||||
Studied Phenotype |
Melanoma [ICD-11:2C30] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.03E-05; Fold-change:-0.440875689; Z-score:-1.692068608 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Multilayered rosettes embryonal tumour |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC12A5 in multilayered rosettes embryonal tumour than that in healthy individual | ||||
Studied Phenotype |
Multilayered rosettes embryonal tumour [ICD-11:2A00.1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:1.50E-13; Fold-change:-0.558340343; Z-score:-2.301500053 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
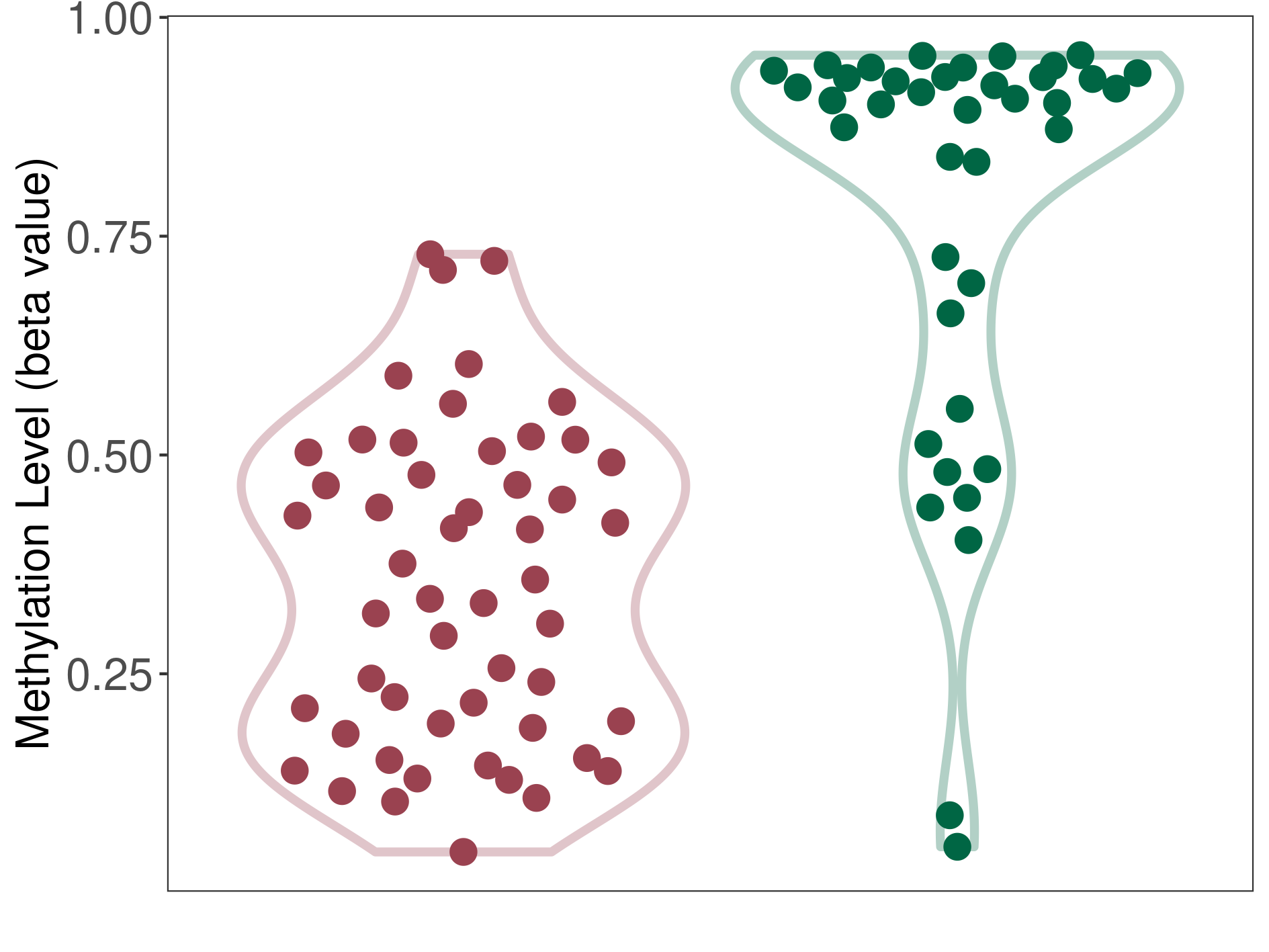
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Third ventricle chordoid glioma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of SLC12A5 in third ventricle chordoid glioma than that in healthy individual | ||||
Studied Phenotype |
Third ventricle chordoid glioma [ICD-11:2A00.0Y] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.005064491; Fold-change:-0.372595446; Z-score:-7.541705034 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
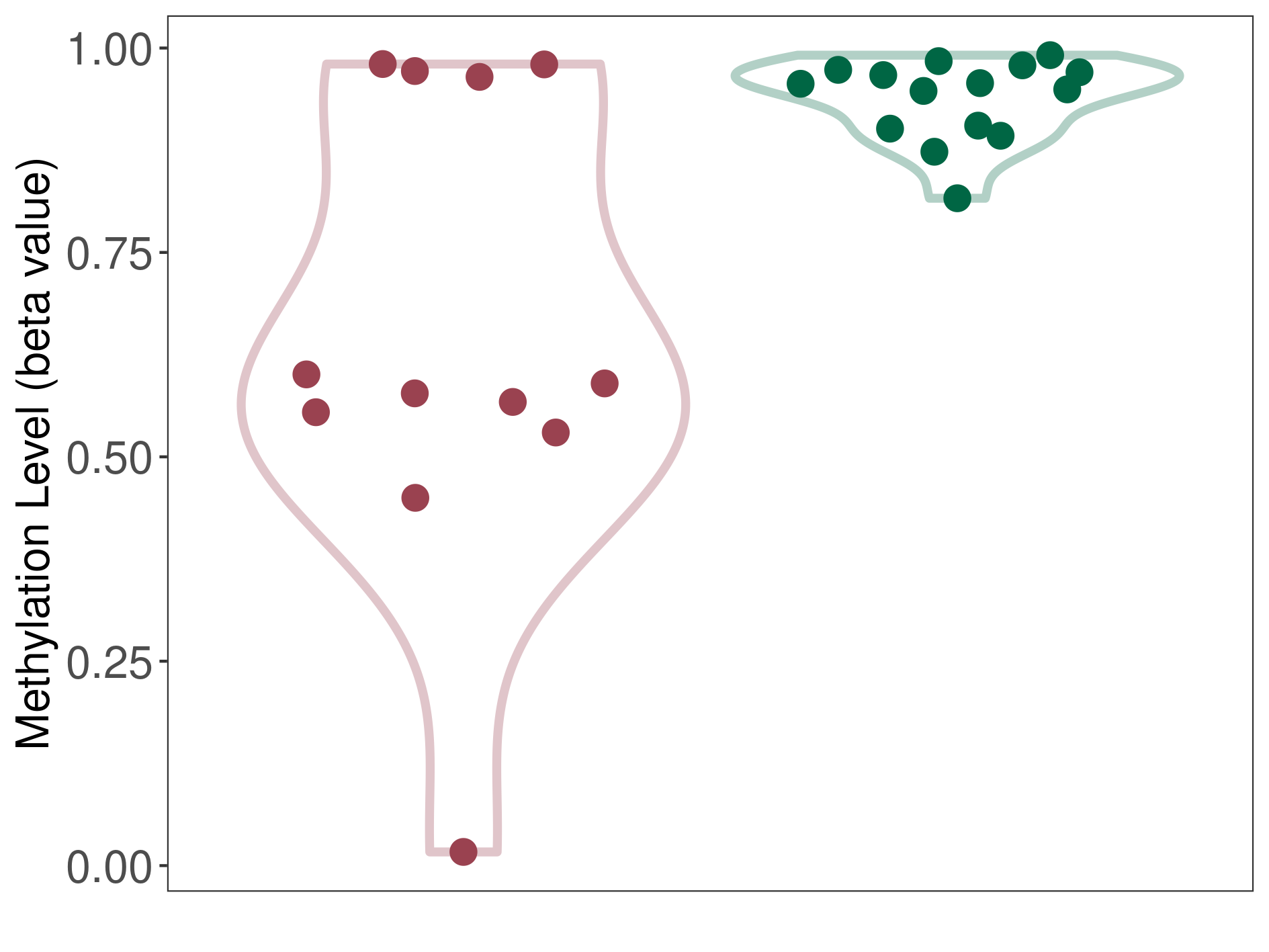
|
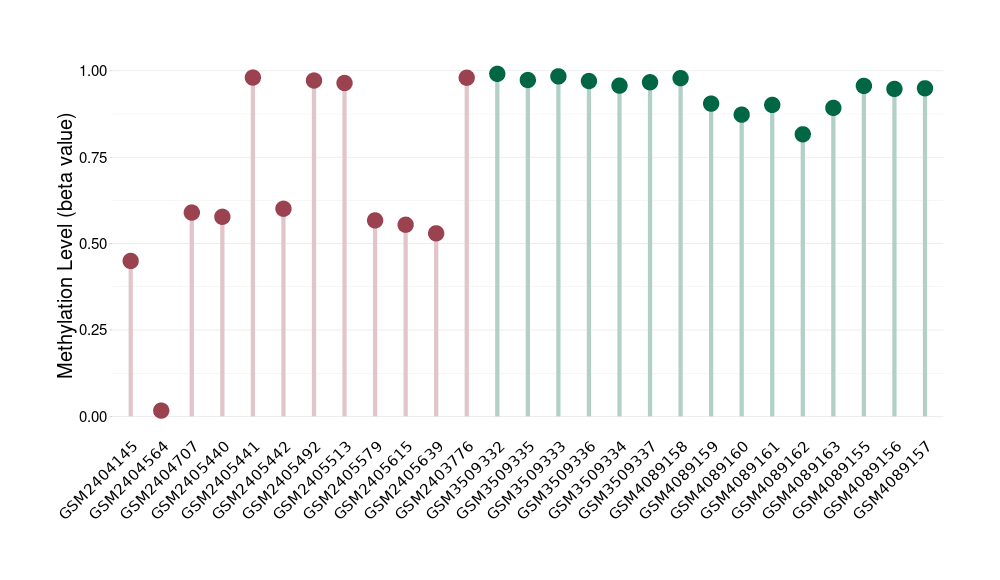 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Gastric cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypermethylation of SLC12A5 in gastric cancer than that in adjacent tissue | ||||
Studied Phenotype |
Gastric cancer [ICD-11:2B72] | ||||
The Methylation Level of Disease Section Compare with the Adjacent Tissue |
p-value:0.005109813; Fold-change:0.240848646; Z-score:7.982504387 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue adjacent to the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
Histone acetylation |
|||||
|
Bone cancer pain |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Hypoacetylation of Slc12a5 in bone cancer pain (compare with normal rats) | [ 16 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes | Down regulation ofSlc12a5 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Bone cancer pain[ ICD-11:MG30.1] | ||||
|
Experimental Material |
Model organism in vivo (mouse) | ||||
|
Additional Notes |
Suppression of HDAC2 in spinal cord alleviates mechanical hyperalgesia and restores KCC2 expression in a rat model of bone cancer pain. | ||||
|
Inflammatory pain |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Hypoacetylation of Slc12a5 in inflammatory pain (compare with normal rats) | [ 17 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes | Down regulation ofSlc12a5 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Inflammatory pain[ ICD-11:MG30.42] | ||||
|
Experimental Material |
Model organism in vivo (mouse) | ||||
|
Additional Notes |
Promoter histone acetylation plays a role in complete Freund's adjuvantinduced suppression of SLC12A5 in the spinal dorsal horn. | ||||
|
Epigenetic Phenomenon2 |
Hypoacetylation of Slc12a5 in inflammatory pain (compare with normal rats) | [ 17 ] | |||
|
Location |
Promoter | ||||
|
Epigenetic Type |
Histone acetylation | Experiment Method | Chromatin immunoprecipitation | ||
|
Related Molecular Changes | Down regulation ofSlc12a5 | Experiment Method | Western Blot | ||
|
Studied Phenotype |
Inflammatory pain[ ICD-11:MG30.42] | ||||
|
Experimental Material |
Model organism in vivo (mouse) | ||||
|
Additional Notes |
Promoter histone acetylation plays a role in complete Freund's adjuvantinduced suppression of SLC12A5 in the spinal dorsal horn. | ||||
|
microRNA |
|||||
|
Unclear Phenotype |
38 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
miR-1237 directly targets SLC12A5 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1237 | miRNA Mature ID | miR-1237-5p | ||
|
miRNA Sequence |
CGGGGGCGGGGCCGAAGCGCG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon2 |
miR-203a directly targets SLC12A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-203a | miRNA Mature ID | miR-203a-3p | ||
|
miRNA Sequence |
GUGAAAUGUUUAGGACCACUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Epigenetic Phenomenon3 |
miR-224 directly targets SLC12A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-224 | miRNA Mature ID | miR-224-5p | ||
|
miRNA Sequence |
UCAAGUCACUAGUGGUUCCGUUUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Epigenetic Phenomenon4 |
miR-2277 directly targets SLC12A5 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-2277 | miRNA Mature ID | miR-2277-3p | ||
|
miRNA Sequence |
UGACAGCGCCCUGCCUGGCUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon5 |
miR-25 directly targets SLC12A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-25 | miRNA Mature ID | miR-25-3p | ||
|
miRNA Sequence |
CAUUGCACUUGUCUCGGUCUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Epigenetic Phenomenon6 |
miR-3178 directly targets SLC12A5 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3178 | miRNA Mature ID | miR-3178 | ||
|
miRNA Sequence |
GGGGCGCGGCCGGAUCG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon7 |
miR-32 directly targets SLC12A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-32 | miRNA Mature ID | miR-32-5p | ||
|
miRNA Sequence |
UAUUGCACAUUACUAAGUUGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Epigenetic Phenomenon8 |
miR-328 directly targets SLC12A5 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-328 | miRNA Mature ID | miR-328-5p | ||
|
miRNA Sequence |
GGGGGGGCAGGAGGGGCUCAGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon9 |
miR-3614 directly targets SLC12A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3614 | miRNA Mature ID | miR-3614-5p | ||
|
miRNA Sequence |
CCACUUGGAUCUGAAGGCUGCCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon10 |
miR-363 directly targets SLC12A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-363 | miRNA Mature ID | miR-363-3p | ||
|
miRNA Sequence |
AAUUGCACGGUAUCCAUCUGUA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Epigenetic Phenomenon11 |
miR-367 directly targets SLC12A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-367 | miRNA Mature ID | miR-367-3p | ||
|
miRNA Sequence |
AAUUGCACUUUAGCAAUGGUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Epigenetic Phenomenon12 |
miR-4285 directly targets SLC12A5 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4285 | miRNA Mature ID | miR-4285 | ||
|
miRNA Sequence |
GCGGCGAGUCCGACUCAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon13 |
miR-4313 directly targets SLC12A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4313 | miRNA Mature ID | miR-4313 | ||
|
miRNA Sequence |
AGCCCCCUGGCCCCAAACCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon14 |
miR-4325 directly targets SLC12A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4325 | miRNA Mature ID | miR-4325 | ||
|
miRNA Sequence |
UUGCACUUGUCUCAGUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Epigenetic Phenomenon15 |
miR-433 directly targets SLC12A5 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-433 | miRNA Mature ID | miR-433-5p | ||
|
miRNA Sequence |
UACGGUGAGCCUGUCAUUAUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon16 |
miR-4433a directly targets SLC12A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4433a | miRNA Mature ID | miR-4433a-5p | ||
|
miRNA Sequence |
CGUCCCACCCCCCACUCCUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon17 |
miR-4433b directly targets SLC12A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4433b | miRNA Mature ID | miR-4433b-5p | ||
|
miRNA Sequence |
AUGUCCCACCCCCACUCCUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon18 |
miR-4469 directly targets SLC12A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4469 | miRNA Mature ID | miR-4469 | ||
|
miRNA Sequence |
GCUCCCUCUAGGGUCGCUCGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon19 |
miR-4479 directly targets SLC12A5 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4479 | miRNA Mature ID | miR-4479 | ||
|
miRNA Sequence |
CGCGCGGCCGUGCUCGGAGCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon20 |
miR-4488 directly targets SLC12A5 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4488 | miRNA Mature ID | miR-4488 | ||
|
miRNA Sequence |
AGGGGGCGGGCUCCGGCG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon21 |
miR-4646 directly targets SLC12A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4646 | miRNA Mature ID | miR-4646-3p | ||
|
miRNA Sequence |
AUUGUCCCUCUCCCUUCCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon22 |
miR-4667 directly targets SLC12A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4667 | miRNA Mature ID | miR-4667-3p | ||
|
miRNA Sequence |
UCCCUCCUUCUGUCCCCACAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon23 |
miR-4697 directly targets SLC12A5 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4697 | miRNA Mature ID | miR-4697-5p | ||
|
miRNA Sequence |
AGGGGGCGCAGUCACUGACGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon24 |
miR-4726 directly targets SLC12A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4726 | miRNA Mature ID | miR-4726-3p | ||
|
miRNA Sequence |
ACCCAGGUUCCCUCUGGCCGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon25 |
miR-5193 directly targets SLC12A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5193 | miRNA Mature ID | miR-5193 | ||
|
miRNA Sequence |
UCCUCCUCUACCUCAUCCCAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon26 |
miR-542 directly targets SLC12A5 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-542 | miRNA Mature ID | miR-542-3p | ||
|
miRNA Sequence |
UGUGACAGAUUGAUAACUGAAA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon27 |
miR-548az directly targets SLC12A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-548az | miRNA Mature ID | miR-548az-5p | ||
|
miRNA Sequence |
CAAAAGUGAUUGUGGUUUUUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Epigenetic Phenomenon28 |
miR-548n directly targets SLC12A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-548n | miRNA Mature ID | miR-548n | ||
|
miRNA Sequence |
CAAAAGUAAUUGUGGAUUUUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Epigenetic Phenomenon29 |
miR-548t directly targets SLC12A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-548t | miRNA Mature ID | miR-548t-5p | ||
|
miRNA Sequence |
CAAAAGUGAUCGUGGUUUUUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Epigenetic Phenomenon30 |
miR-6075 directly targets SLC12A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6075 | miRNA Mature ID | miR-6075 | ||
|
miRNA Sequence |
ACGGCCCAGGCGGCAUUGGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon31 |
miR-6500 directly targets SLC12A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6500 | miRNA Mature ID | miR-6500-3p | ||
|
miRNA Sequence |
ACACUUGUUGGGAUGACCUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon32 |
miR-6749 directly targets SLC12A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6749 | miRNA Mature ID | miR-6749-3p | ||
|
miRNA Sequence |
CUCCUCCCCUGCCUGGCCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon33 |
miR-6840 directly targets SLC12A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6840 | miRNA Mature ID | miR-6840-3p | ||
|
miRNA Sequence |
GCCCAGGACUUUGUGCGGGGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon34 |
miR-6885 directly targets SLC12A5 | [ 18 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6885 | miRNA Mature ID | miR-6885-5p | ||
|
miRNA Sequence |
AGGGGGGCACUGCGCAAGCAAAGCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon35 |
miR-6887 directly targets SLC12A5 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6887 | miRNA Mature ID | miR-6887-3p | ||
|
miRNA Sequence |
UCCCCUCCACUUUCCUCCUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon36 |
miR-7703 directly targets SLC12A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-7703 | miRNA Mature ID | miR-7703 | ||
|
miRNA Sequence |
UUGCACUCUGGCCUUCUCCCAGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Epigenetic Phenomenon37 |
miR-92a directly targets SLC12A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-92a | miRNA Mature ID | miR-92a-3p | ||
|
miRNA Sequence |
UAUUGCACUUGUCCCGGCCUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
|
Epigenetic Phenomenon38 |
miR-92b directly targets SLC12A5 | [ 19 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-92b | miRNA Mature ID | miR-92b-3p | ||
|
miRNA Sequence |
UAUUGCACUCGUCCCGGCCUCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human breast adenocarcinoma cell line (MCF-7) | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
