Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0051 Transporter Info | ||||
| Gene Name | ABCB5 | ||||
| Transporter Name | ATP-binding cassette sub-family B member 5 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Atypical teratoid rhabdoid tumor |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of ABCB5 in atypical teratoid rhabdoid tumor | [ 1 ] | |||
|
Location |
5'UTR (cg21202529) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:5.13E-06; Z-score:-7.76E-01 | ||
|
Methylation in Case |
8.72E-01 (Median) | Methylation in Control | 8.96E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of ABCB5 in bladder cancer | [ 2 ] | |||
|
Location |
5'UTR (cg21202529) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.28E+00 | Statistic Test | p-value:2.02E-03; Z-score:-2.73E+00 | ||
|
Methylation in Case |
6.75E-01 (Median) | Methylation in Control | 8.66E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of ABCB5 in bladder cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg20966504) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.54E+00 | Statistic Test | p-value:1.36E-07; Z-score:-1.44E+01 | ||
|
Methylation in Case |
5.50E-01 (Median) | Methylation in Control | 8.49E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of ABCB5 in bladder cancer | [ 2 ] | |||
|
Location |
TSS1500 (cg08888968) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:5.20E-03; Z-score:-2.66E+00 | ||
|
Methylation in Case |
8.94E-01 (Median) | Methylation in Control | 9.19E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of ABCB5 in bladder cancer | [ 2 ] | |||
|
Location |
TSS200 (cg14878128) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.81E+00 | Statistic Test | p-value:2.58E-03; Z-score:-2.40E+00 | ||
|
Methylation in Case |
4.05E-01 (Median) | Methylation in Control | 7.32E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of ABCB5 in breast cancer | [ 3 ] | |||
|
Location |
5'UTR (cg21202529) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.27E-03; Z-score:-2.21E-01 | ||
|
Methylation in Case |
8.76E-01 (Median) | Methylation in Control | 8.83E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of ABCB5 in breast cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg20966504) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:2.85E-07; Z-score:1.64E+00 | ||
|
Methylation in Case |
7.63E-01 (Median) | Methylation in Control | 6.44E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of ABCB5 in breast cancer | [ 3 ] | |||
|
Location |
TSS1500 (cg08888968) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:3.06E-02; Z-score:-3.33E-01 | ||
|
Methylation in Case |
8.98E-01 (Median) | Methylation in Control | 9.11E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of ABCB5 in breast cancer | [ 3 ] | |||
|
Location |
TSS200 (cg14878128) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:2.59E-02; Z-score:5.34E-01 | ||
|
Methylation in Case |
5.70E-01 (Median) | Methylation in Control | 5.17E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
5 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of ABCB5 in colorectal cancer | [ 4 ] | |||
|
Location |
5'UTR (cg21202529) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:2.47E-02; Z-score:-6.03E-01 | ||
|
Methylation in Case |
7.86E-01 (Median) | Methylation in Control | 8.25E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of ABCB5 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg20966504) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.41E-06; Z-score:-2.46E+00 | ||
|
Methylation in Case |
8.32E-01 (Median) | Methylation in Control | 8.81E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of ABCB5 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS1500 (cg08888968) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.37E-04; Z-score:-8.45E-01 | ||
|
Methylation in Case |
9.52E-01 (Median) | Methylation in Control | 9.58E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of ABCB5 in colorectal cancer | [ 4 ] | |||
|
Location |
TSS200 (cg14878128) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.19E+00 | Statistic Test | p-value:9.82E-05; Z-score:-1.46E+00 | ||
|
Methylation in Case |
6.41E-01 (Median) | Methylation in Control | 7.65E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of ABCB5 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
5'UTR (cg21202529) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.28E+00 | Statistic Test | p-value:1.06E-08; Z-score:-2.27E+00 | ||
|
Methylation in Case |
6.13E-01 (Median) | Methylation in Control | 7.83E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of ABCB5 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg08888968) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.15E+00 | Statistic Test | p-value:2.45E-09; Z-score:-3.23E+00 | ||
|
Methylation in Case |
7.57E-01 (Median) | Methylation in Control | 8.67E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of ABCB5 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg14878128) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.45E+00 | Statistic Test | p-value:4.79E-07; Z-score:-1.36E+00 | ||
|
Methylation in Case |
3.63E-01 (Median) | Methylation in Control | 5.26E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
HIV infection |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of ABCB5 in HIV infection | [ 6 ] | |||
|
Location |
5'UTR (cg21202529) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:3.87E-02; Z-score:3.92E-01 | ||
|
Methylation in Case |
9.51E-01 (Median) | Methylation in Control | 9.43E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Pancretic ductal adenocarcinoma |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of ABCB5 in pancretic ductal adenocarcinoma | [ 7 ] | |||
|
Location |
5'UTR (cg19300741) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.56E+00 | Statistic Test | p-value:4.99E-18; Z-score:2.96E+00 | ||
|
Methylation in Case |
2.96E-01 (Median) | Methylation in Control | 1.90E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of ABCB5 in pancretic ductal adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg24151995) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.10E+00 | Statistic Test | p-value:4.46E-04; Z-score:6.95E-01 | ||
|
Methylation in Case |
3.15E-01 (Median) | Methylation in Control | 2.87E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of ABCB5 in pancretic ductal adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg23244463) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:9.61E-03; Z-score:-6.03E-02 | ||
|
Methylation in Case |
8.07E-01 (Median) | Methylation in Control | 8.10E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of ABCB5 in pancretic ductal adenocarcinoma | [ 7 ] | |||
|
Location |
Body (cg16793755) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.08E+00 | Statistic Test | p-value:1.26E-02; Z-score:6.60E-01 | ||
|
Methylation in Case |
7.47E-01 (Median) | Methylation in Control | 6.94E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Systemic lupus erythematosus |
2 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of ABCB5 in systemic lupus erythematosus | [ 8 ] | |||
|
Location |
5'UTR (cg21202529) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.00E+00 | Statistic Test | p-value:3.65E-02; Z-score:-1.52E-01 | ||
|
Methylation in Case |
9.34E-01 (Median) | Methylation in Control | 9.37E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of ABCB5 in systemic lupus erythematosus | [ 8 ] | |||
|
Location |
TSS1500 (cg20966504) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:8.28E-03; Z-score:-2.96E-01 | ||
|
Methylation in Case |
9.07E-01 (Median) | Methylation in Control | 9.14E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of ABCB5 in panic disorder | [ 9 ] | |||
|
Location |
TSS1500 (cg20966504) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:8.73E-03; Z-score:-1.91E-01 | ||
|
Methylation in Case |
2.61E+00 (Median) | Methylation in Control | 2.68E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of ABCB5 in papillary thyroid cancer | [ 10 ] | |||
|
Location |
TSS200 (cg14878128) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:4.19E-03; Z-score:-5.96E-01 | ||
|
Methylation in Case |
7.02E-01 (Median) | Methylation in Control | 7.52E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Esthesioneuroblastoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypomethylation of ABCB5 in esthesioneuroblastoma than that in healthy individual | ||||
Studied Phenotype |
Esthesioneuroblastoma [ICD-11:2D50.1] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:2.97E-06; Fold-change:-0.231921153; Z-score:-1.8149088 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
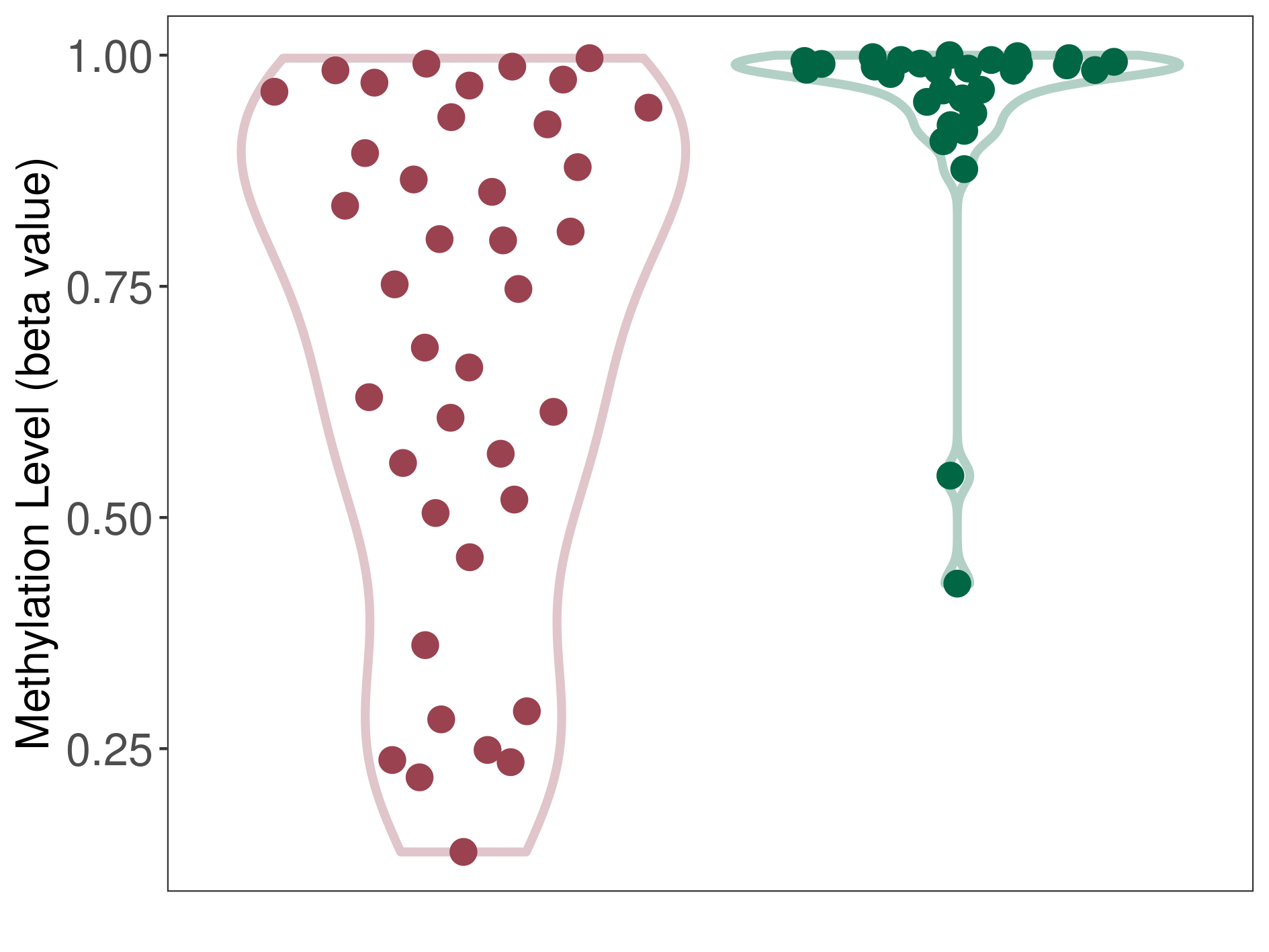
|
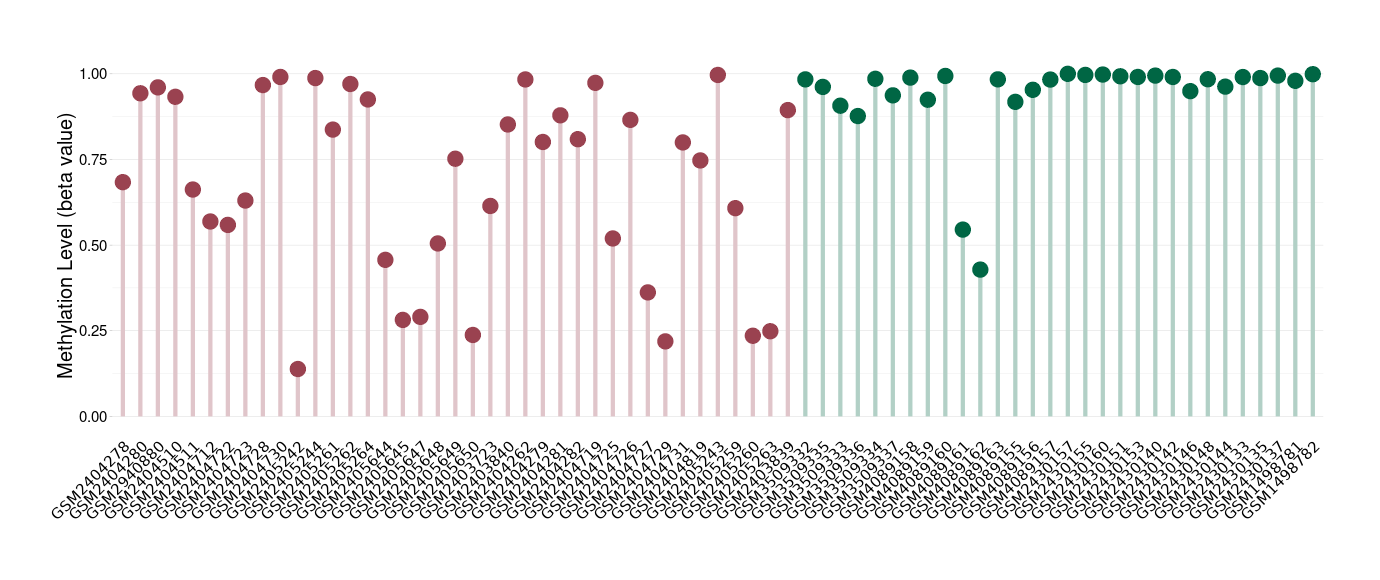 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
Melanocytoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Significant hypomethylation of ABCB5 in melanocytoma than that in healthy individual | ||||
Studied Phenotype |
Melanocytoma [ICD-11:2F36.2] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:2.84E-16; Fold-change:-0.821669195; Z-score:-31.2495816 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
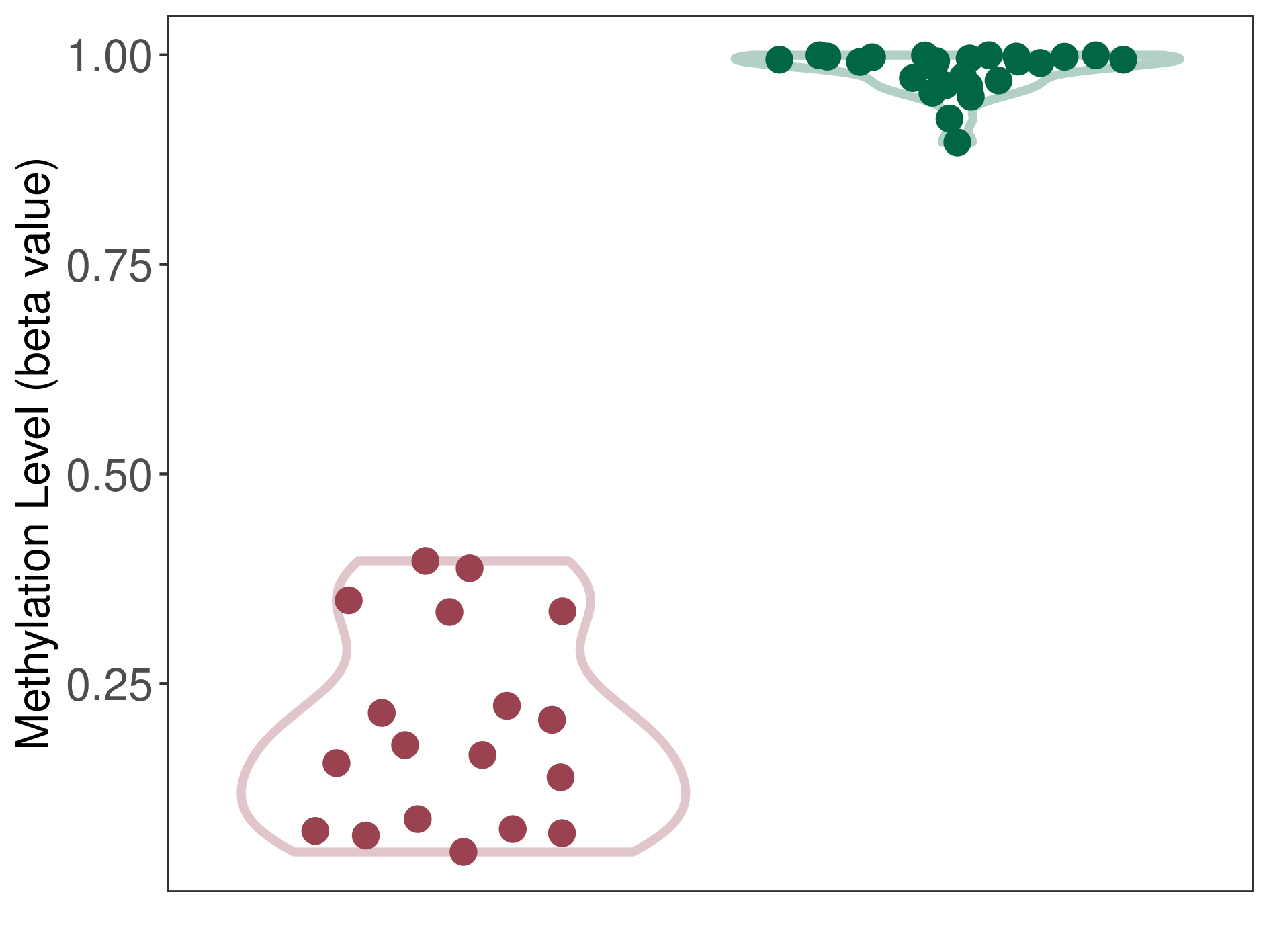
|
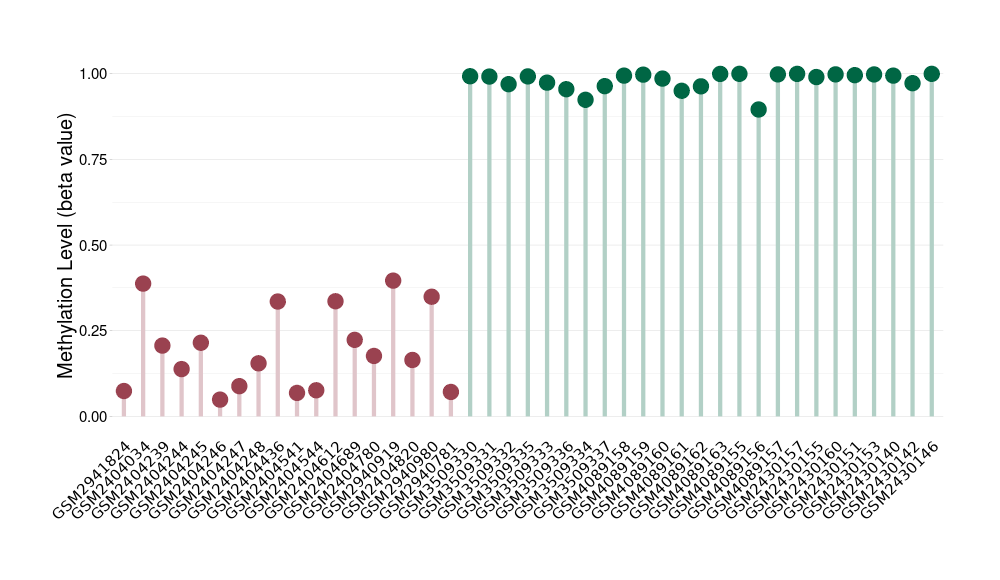 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
microRNA |
|||||
|
Melanoma |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Higher expression of miR-340-5p in melanoma (compare with hypoxic conditions counterpart cells) | [ 11 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | . | ||
|
Related Molecular Changes | Down regulation ofABCB5 | Experiment Method | RT-qPCR | ||
|
miRNA Stemloop ID |
miR-340 | miRNA Mature ID | miR-340-5p | ||
|
miRNA Sequence |
UUAUAAAGCAAUGAGACUGAUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Studied Phenotype |
Melanoma[ ICD-11:2C30] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Additional Notes |
Diminution of miR-340-5p levels is responsible for increased expression of ABCB5 in melanoma cells under oxygen-deprived conditions. | ||||
|
Colon cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Higher expression of miR-522 in colon cancer (compare with doxorubicin-resistance cells) | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Luciferase reporter assay | ||
|
Related Molecular Changes | Down regulation ofABCB5 | Experiment Method | RT-qPCR | ||
|
miRNA Stemloop ID |
miR-522 | miRNA Mature ID | miR-522-5p | ||
|
miRNA Sequence |
CUCUAGAGGGAAGCGCUUUCUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Human colon cancer cell line (HT-29) | ||||
|
Additional Notes |
MicroRNA-522 reverses drug resistance of doxorubicin-induced HT29 colon cancer cell by targeting ABCB5. | ||||
|
Unclear Phenotype |
28 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
miR-1 directly targets ABCB5 | [ 13 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Microarray | ||
|
miRNA Stemloop ID |
miR-1 | miRNA Mature ID | miR-1-3p | ||
|
miRNA Sequence |
UGGAAUGUAAAGAAGUAUGUAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human cervical cancer cell line (Hela) | ||||
|
Epigenetic Phenomenon2 |
miR-100 directly targets ABCB5 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-100 | miRNA Mature ID | miR-100-3p | ||
|
miRNA Sequence |
CAAGCUUGUAUCUAUAGGUAUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon3 |
miR-105 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-105 | miRNA Mature ID | miR-105-5p | ||
|
miRNA Sequence |
UCAAAUGCUCAGACUCCUGUGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon4 |
miR-1304 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-1304 | miRNA Mature ID | miR-1304-3p | ||
|
miRNA Sequence |
UCUCACUGUAGCCUCGAACCCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon5 |
miR-1307 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-1307 | miRNA Mature ID | miR-1307-3p | ||
|
miRNA Sequence |
ACUCGGCGUGGCGUCGGUCGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon6 |
miR-192 directly targets ABCB5 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-192 | miRNA Mature ID | miR-192-3p | ||
|
miRNA Sequence |
CUGCCAAUUCCAUAGGUCACAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon7 |
miR-330 directly targets ABCB5 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-330 | miRNA Mature ID | miR-330-3p | ||
|
miRNA Sequence |
GCAAAGCACACGGCCUGCAGAGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon8 |
miR-3671 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-3671 | miRNA Mature ID | miR-3671 | ||
|
miRNA Sequence |
AUCAAAUAAGGACUAGUCUGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon9 |
miR-4284 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4284 | miRNA Mature ID | miR-4284 | ||
|
miRNA Sequence |
GGGCUCACAUCACCCCAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon10 |
miR-4311 directly targets ABCB5 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4311 | miRNA Mature ID | miR-4311 | ||
|
miRNA Sequence |
GAAAGAGAGCUGAGUGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon11 |
miR-4480 directly targets ABCB5 | [ 14 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4480 | miRNA Mature ID | miR-4480 | ||
|
miRNA Sequence |
AGCCAAGUGGAAGUUACUUUA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon12 |
miR-4638 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4638 | miRNA Mature ID | miR-4638-5p | ||
|
miRNA Sequence |
ACUCGGCUGCGGUGGACAAGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon13 |
miR-4676 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4676 | miRNA Mature ID | miR-4676-5p | ||
|
miRNA Sequence |
GAGCCAGUGGUGAGACAGUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon14 |
miR-4710 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4710 | miRNA Mature ID | miR-4710 | ||
|
miRNA Sequence |
GGGUGAGGGCAGGUGGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon15 |
miR-4772 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-4772 | miRNA Mature ID | miR-4772-3p | ||
|
miRNA Sequence |
CCUGCAACUUUGCCUGAUCAGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon16 |
miR-5001 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5001 | miRNA Mature ID | miR-5001-3p | ||
|
miRNA Sequence |
UUCUGCCUCUGUCCAGGUCCUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon17 |
miR-522 directly targets ABCB5 | [ 12 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Flow//GFP reporter assay//qRT-PCR//Western blot | ||
|
miRNA Stemloop ID |
miR-522 | miRNA Mature ID | miR-522-5p | ||
|
miRNA Sequence |
CUCUAGAGGGAAGCGCUUUCUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human colon cancer cell line (HT-29) | ||||
|
Epigenetic Phenomenon18 |
miR-5571 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-5571 | miRNA Mature ID | miR-5571-3p | ||
|
miRNA Sequence |
GUCCUAGGAGGCUCCUCUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon19 |
miR-580 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-580 | miRNA Mature ID | miR-580-5p | ||
|
miRNA Sequence |
UAAUGAUUCAUCAGACUCAGAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon20 |
miR-607 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-607 | miRNA Mature ID | miR-607 | ||
|
miRNA Sequence |
GUUCAAAUCCAGAUCUAUAAC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon21 |
miR-6741 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6741 | miRNA Mature ID | miR-6741-3p | ||
|
miRNA Sequence |
UCGGCUCUCUCCCUCACCCUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon22 |
miR-6778 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6778 | miRNA Mature ID | miR-6778-3p | ||
|
miRNA Sequence |
UGCCUCCCUGACAUUCCACAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon23 |
miR-6791 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6791 | miRNA Mature ID | miR-6791-3p | ||
|
miRNA Sequence |
UGCCUCCUUGGUCUCCGGCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon24 |
miR-6829 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6829 | miRNA Mature ID | miR-6829-3p | ||
|
miRNA Sequence |
UGCCUCCUCCGUGGCCUCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon25 |
miR-6836 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6836 | miRNA Mature ID | miR-6836-3p | ||
|
miRNA Sequence |
AUGCCUCCCCCGGCCCCGCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon26 |
miR-6890 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-6890 | miRNA Mature ID | miR-6890-3p | ||
|
miRNA Sequence |
CCACUGCCUAUGCCCCACAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon27 |
miR-7853 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-7853 | miRNA Mature ID | miR-7853-5p | ||
|
miRNA Sequence |
UCAAAUGCAGAUCCUGACUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon28 |
miR-7855 directly targets ABCB5 | [ 15 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | HITS-CLIP | ||
|
miRNA Stemloop ID |
miR-7855 | miRNA Mature ID | miR-7855-5p | ||
|
miRNA Sequence |
UUGGUGAGGACCCCAAGCUCGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
