Detail Information of Epigenetic Regulations
| General Information of Drug Transporter (DT) | |||||
|---|---|---|---|---|---|
| DT ID | DTD0007 Transporter Info | ||||
| Gene Name | SLC47A1 | ||||
| Transporter Name | Multidrug and toxin extrusion protein 1 | ||||
| Gene ID | |||||
| UniProt ID | |||||
| Epigenetic Regulations of This DT (EGR) | |||||
|---|---|---|---|---|---|
|
Methylation |
|||||
|
Diabetes |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Hypermethylation of SLC47A1 in diabetes | [ 1 ] | |||
|
Location |
Promoter (11 CpG sites) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | HumanMethylation450 BeadChip | ||
|
Related Molecular Changes | Down regulation ofSLC47A1 | Experiment Method | BeadChip | ||
|
Studied Phenotype |
Diabetes[ ICD-11:5A10-5A14] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Additional Notes |
Lower average and promoter DNA methylation of SLC47A1 was found in diabetic subjects receiving just metformin,compared to those who took insulin plus metformin or no diabetes medication. | ||||
|
Human liver tissue |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Hypermethylation of SLC47A1 in human liver tissue | [ 2 ] | |||
|
Location |
27 kb upstream of gene | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Bisulfite sequencing | ||
|
Related Molecular Changes | Down regulation ofSLC47A1 | Experiment Method | Semiquantitative RT-PCR | ||
|
Studied Phenotype |
Human liver tissue | ||||
|
Experimental Material |
Caucasian patient tissue samples | ||||
|
Additional Notes |
A relationship was not found between DNA methylation in the SLC47A1 promoter and MATE1 mRNA expression. | ||||
|
Non-alcoholic fatty liver disease |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Hypermethylation of SLC47A1 in non-alcoholic fatty liver disease | [ 3 ] | |||
|
Location |
6 CpG sites | ||||
|
Epigenetic Type |
Methylation | Experiment Method | HumanMethylation450 BeadChip | ||
|
Related Molecular Changes | Down regulation ofSLC47A1 | Experiment Method | Methylation microarray | ||
|
Studied Phenotype |
Non-alcoholic fatty liver disease[ ICD-11:DB92] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Additional Notes |
SLC47A1 was detected to be significantly inversely associated with the transcriptional expression. | ||||
|
Colon cancer |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC47A1 in colon adenocarcinoma | [ 4 ] | |||
|
Location |
5'UTR (cg05373457) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.30E+00 | Statistic Test | p-value:1.89E-06; Z-score:2.08E+00 | ||
|
Methylation in Case |
5.58E-01 (Median) | Methylation in Control | 4.29E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC47A1 in colon adenocarcinoma | [ 4 ] | |||
|
Location |
TSS1500 (cg02577240) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.16E+00 | Statistic Test | p-value:7.18E-04; Z-score:-1.49E+00 | ||
|
Methylation in Case |
6.01E-01 (Median) | Methylation in Control | 6.98E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC47A1 in colon adenocarcinoma | [ 4 ] | |||
|
Location |
TSS200 (cg05311412) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.34E+00 | Statistic Test | p-value:7.92E-07; Z-score:3.02E+00 | ||
|
Methylation in Case |
6.11E-01 (Median) | Methylation in Control | 2.60E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC47A1 in colon adenocarcinoma | [ 4 ] | |||
|
Location |
Body (cg01454305) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.46E+00 | Statistic Test | p-value:3.53E-07; Z-score:-3.88E+00 | ||
|
Methylation in Case |
4.91E-01 (Median) | Methylation in Control | 7.16E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC47A1 in colon adenocarcinoma | [ 4 ] | |||
|
Location |
Body (cg10311318) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:4.42E-04; Z-score:8.90E-01 | ||
|
Methylation in Case |
5.39E-01 (Median) | Methylation in Control | 4.72E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC47A1 in colon adenocarcinoma | [ 4 ] | |||
|
Location |
Body (cg12866656) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.19E+00 | Statistic Test | p-value:2.22E-03; Z-score:-1.58E+00 | ||
|
Methylation in Case |
5.17E-01 (Median) | Methylation in Control | 6.16E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC47A1 in colon adenocarcinoma | [ 4 ] | |||
|
Location |
3'UTR (cg08828816) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:4.55E-03; Z-score:-2.33E+00 | ||
|
Methylation in Case |
6.88E-01 (Median) | Methylation in Control | 7.42E-01 (Median) | ||
|
Studied Phenotype |
Colon cancer[ ICD-11:2B90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Hepatocellular carcinoma |
18 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
5'UTR (cg14932645) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:5.28E-10; Z-score:-3.98E+00 | ||
|
Methylation in Case |
7.68E-01 (Median) | Methylation in Control | 8.67E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg21692194) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.17E+00 | Statistic Test | p-value:8.19E-06; Z-score:-6.73E-01 | ||
|
Methylation in Case |
1.09E-01 (Median) | Methylation in Control | 1.28E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg15971010) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.40E+00 | Statistic Test | p-value:6.81E-04; Z-score:-8.39E-01 | ||
|
Methylation in Case |
1.08E-01 (Median) | Methylation in Control | 1.50E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg25387636) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.46E+00 | Statistic Test | p-value:1.62E-02; Z-score:-8.51E-01 | ||
|
Methylation in Case |
5.88E-02 (Median) | Methylation in Control | 8.59E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS1500 (cg12133118) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:1.66E-02; Z-score:-7.27E-01 | ||
|
Methylation in Case |
3.22E-01 (Median) | Methylation in Control | 3.67E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg07829432) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.24E+00 | Statistic Test | p-value:6.45E-04; Z-score:-7.82E-01 | ||
|
Methylation in Case |
8.02E-02 (Median) | Methylation in Control | 9.95E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg15014549) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.13E+00 | Statistic Test | p-value:6.52E-03; Z-score:-5.87E-01 | ||
|
Methylation in Case |
6.69E-02 (Median) | Methylation in Control | 7.54E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
TSS200 (cg13004927) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.28E+00 | Statistic Test | p-value:1.84E-02; Z-score:-2.52E-01 | ||
|
Methylation in Case |
2.58E-02 (Median) | Methylation in Control | 3.31E-02 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg02068832) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.79E+00 | Statistic Test | p-value:1.39E-18; Z-score:-6.05E+00 | ||
|
Methylation in Case |
4.16E-01 (Median) | Methylation in Control | 7.44E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg16925177) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.67E+00 | Statistic Test | p-value:3.76E-17; Z-score:-5.57E+00 | ||
|
Methylation in Case |
4.81E-01 (Median) | Methylation in Control | 8.03E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg05510976) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.51E+00 | Statistic Test | p-value:2.88E-16; Z-score:-6.93E+00 | ||
|
Methylation in Case |
5.25E-01 (Median) | Methylation in Control | 7.91E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg07566700) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.36E+00 | Statistic Test | p-value:5.37E-16; Z-score:-4.42E+00 | ||
|
Methylation in Case |
5.62E-01 (Median) | Methylation in Control | 7.66E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg00601711) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:4.46E-12; Z-score:2.14E+00 | ||
|
Methylation in Case |
7.63E-01 (Median) | Methylation in Control | 6.42E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg10718608) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.32E+00 | Statistic Test | p-value:4.40E-08; Z-score:-1.60E+00 | ||
|
Methylation in Case |
3.71E-01 (Median) | Methylation in Control | 4.91E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg08895056) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.31E+00 | Statistic Test | p-value:8.41E-08; Z-score:-1.61E+00 | ||
|
Methylation in Case |
3.71E-01 (Median) | Methylation in Control | 4.87E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg20930201) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.27E+00 | Statistic Test | p-value:1.63E-05; Z-score:-7.83E-01 | ||
|
Methylation in Case |
1.15E-01 (Median) | Methylation in Control | 1.45E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
Body (cg12894055) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.38E+00 | Statistic Test | p-value:5.37E-03; Z-score:-1.06E+00 | ||
|
Methylation in Case |
9.94E-02 (Median) | Methylation in Control | 1.37E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon18 |
Methylation of SLC47A1 in hepatocellular carcinoma | [ 5 ] | |||
|
Location |
3'UTR (cg12550399) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:7.52E-05; Z-score:3.90E-01 | ||
|
Methylation in Case |
7.69E-01 (Median) | Methylation in Control | 7.40E-01 (Median) | ||
|
Studied Phenotype |
Hepatocellular carcinoma[ ICD-11:2C12.02] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Pancretic ductal adenocarcinoma |
15 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC47A1 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
5'UTR (cg26590537) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.70E+00 | Statistic Test | p-value:1.17E-24; Z-score:3.82E+00 | ||
|
Methylation in Case |
4.11E-01 (Median) | Methylation in Control | 1.52E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC47A1 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg24079038) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.48E+00 | Statistic Test | p-value:5.82E-10; Z-score:-1.78E+00 | ||
|
Methylation in Case |
2.97E-01 (Median) | Methylation in Control | 4.39E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC47A1 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
TSS1500 (cg18646365) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.36E+00 | Statistic Test | p-value:1.57E-08; Z-score:1.31E+00 | ||
|
Methylation in Case |
3.98E-01 (Median) | Methylation in Control | 2.93E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC47A1 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
TSS200 (cg06750832) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:5.47E+00 | Statistic Test | p-value:3.34E-33; Z-score:5.41E+00 | ||
|
Methylation in Case |
4.26E-01 (Median) | Methylation in Control | 7.79E-02 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC47A1 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg14994247) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.21E+00 | Statistic Test | p-value:2.01E-20; Z-score:-2.74E+00 | ||
|
Methylation in Case |
3.94E-01 (Median) | Methylation in Control | 4.76E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC47A1 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg04320476) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.09E+00 | Statistic Test | p-value:7.13E-09; Z-score:-1.61E+00 | ||
|
Methylation in Case |
7.94E-01 (Median) | Methylation in Control | 8.67E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC47A1 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg10605137) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:1.54E-05; Z-score:-9.81E-01 | ||
|
Methylation in Case |
3.28E-01 (Median) | Methylation in Control | 3.61E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC47A1 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg04726373) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:1.65E-05; Z-score:-9.44E-01 | ||
|
Methylation in Case |
7.20E-01 (Median) | Methylation in Control | 7.73E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC47A1 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg19866594) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.23E-05; Z-score:-3.92E-01 | ||
|
Methylation in Case |
8.70E-01 (Median) | Methylation in Control | 8.78E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC47A1 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg13755144) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.02E+00 | Statistic Test | p-value:2.94E-04; Z-score:5.86E-01 | ||
|
Methylation in Case |
8.87E-01 (Median) | Methylation in Control | 8.68E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC47A1 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg00714531) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:1.25E-03; Z-score:6.19E-01 | ||
|
Methylation in Case |
8.82E-01 (Median) | Methylation in Control | 8.47E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC47A1 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg26129353) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:1.62E-02; Z-score:-6.53E-01 | ||
|
Methylation in Case |
7.43E-01 (Median) | Methylation in Control | 8.17E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC47A1 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg13405678) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.03E+00 | Statistic Test | p-value:2.79E-02; Z-score:5.36E-01 | ||
|
Methylation in Case |
5.86E-01 (Median) | Methylation in Control | 5.67E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC47A1 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
Body (cg22288309) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:2.98E-02; Z-score:-1.32E-01 | ||
|
Methylation in Case |
8.00E-01 (Median) | Methylation in Control | 8.06E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC47A1 in pancretic ductal adenocarcinoma | [ 6 ] | |||
|
Location |
3'UTR (cg06566678) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:4.11E-06; Z-score:1.77E+00 | ||
|
Methylation in Case |
8.55E-01 (Median) | Methylation in Control | 8.22E-01 (Median) | ||
|
Studied Phenotype |
Pancretic ductal adenocarcinoma[ ICD-11:2C10.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Bladder cancer |
14 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC47A1 in bladder cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg01530032) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.33E+00 | Statistic Test | p-value:6.81E-07; Z-score:-4.41E+00 | ||
|
Methylation in Case |
4.58E-01 (Median) | Methylation in Control | 6.10E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC47A1 in bladder cancer | [ 7 ] | |||
|
Location |
TSS1500 (cg15971010) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.35E+00 | Statistic Test | p-value:4.06E-02; Z-score:2.06E+00 | ||
|
Methylation in Case |
3.92E-01 (Median) | Methylation in Control | 2.90E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC47A1 in bladder cancer | [ 7 ] | |||
|
Location |
TSS200 (cg13004927) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.67E+00 | Statistic Test | p-value:3.00E-03; Z-score:1.60E+00 | ||
|
Methylation in Case |
5.99E-02 (Median) | Methylation in Control | 3.59E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC47A1 in bladder cancer | [ 7 ] | |||
|
Location |
TSS200 (cg07829432) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.07E+00 | Statistic Test | p-value:3.22E-02; Z-score:4.08E-01 | ||
|
Methylation in Case |
1.18E-01 (Median) | Methylation in Control | 1.10E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC47A1 in bladder cancer | [ 7 ] | |||
|
Location |
TSS200 (cg15014549) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:4.71E-02; Z-score:-7.98E-02 | ||
|
Methylation in Case |
9.52E-02 (Median) | Methylation in Control | 9.62E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC47A1 in bladder cancer | [ 7 ] | |||
|
Location |
Body (cg24151087) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.86E+00 | Statistic Test | p-value:6.51E-16; Z-score:-2.13E+01 | ||
|
Methylation in Case |
1.40E-01 (Median) | Methylation in Control | 4.01E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC47A1 in bladder cancer | [ 7 ] | |||
|
Location |
Body (cg04308167) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.84E+00 | Statistic Test | p-value:1.65E-08; Z-score:-7.26E+00 | ||
|
Methylation in Case |
3.43E-01 (Median) | Methylation in Control | 6.32E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC47A1 in bladder cancer | [ 7 ] | |||
|
Location |
Body (cg12799818) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-3.91E+00 | Statistic Test | p-value:1.22E-07; Z-score:-7.78E+00 | ||
|
Methylation in Case |
8.98E-02 (Median) | Methylation in Control | 3.51E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC47A1 in bladder cancer | [ 7 ] | |||
|
Location |
Body (cg08895056) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.32E+00 | Statistic Test | p-value:4.88E-07; Z-score:-4.70E+00 | ||
|
Methylation in Case |
5.41E-01 (Median) | Methylation in Control | 7.15E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC47A1 in bladder cancer | [ 7 ] | |||
|
Location |
Body (cg10718608) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.35E+00 | Statistic Test | p-value:1.43E-05; Z-score:-5.59E+00 | ||
|
Methylation in Case |
5.02E-01 (Median) | Methylation in Control | 6.75E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC47A1 in bladder cancer | [ 7 ] | |||
|
Location |
Body (cg20930201) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.46E+00 | Statistic Test | p-value:2.90E-04; Z-score:7.75E+00 | ||
|
Methylation in Case |
2.29E-01 (Median) | Methylation in Control | 1.57E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC47A1 in bladder cancer | [ 7 ] | |||
|
Location |
Body (cg26959235) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.44E+00 | Statistic Test | p-value:2.52E-03; Z-score:2.84E+00 | ||
|
Methylation in Case |
1.58E-01 (Median) | Methylation in Control | 1.09E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC47A1 in bladder cancer | [ 7 ] | |||
|
Location |
Body (cg16887170) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:2.74E-03; Z-score:-2.16E+00 | ||
|
Methylation in Case |
7.06E-01 (Median) | Methylation in Control | 7.85E-01 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC47A1 in bladder cancer | [ 7 ] | |||
|
Location |
Body (cg17332016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.27E+00 | Statistic Test | p-value:1.14E-02; Z-score:9.85E-01 | ||
|
Methylation in Case |
5.12E-02 (Median) | Methylation in Control | 4.02E-02 (Median) | ||
|
Studied Phenotype |
Bladder cancer[ ICD-11:2C94] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Breast cancer |
15 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC47A1 in breast cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg15971010) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.35E+00 | Statistic Test | p-value:2.21E-07; Z-score:1.10E+00 | ||
|
Methylation in Case |
2.49E-01 (Median) | Methylation in Control | 1.85E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC47A1 in breast cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg21692194) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:6.56E-06; Z-score:8.36E-01 | ||
|
Methylation in Case |
1.73E-01 (Median) | Methylation in Control | 1.42E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC47A1 in breast cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg25387636) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.39E+00 | Statistic Test | p-value:1.52E-05; Z-score:1.00E+00 | ||
|
Methylation in Case |
1.40E-01 (Median) | Methylation in Control | 1.00E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC47A1 in breast cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg01530032) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.14E+00 | Statistic Test | p-value:2.09E-04; Z-score:-1.21E+00 | ||
|
Methylation in Case |
5.58E-01 (Median) | Methylation in Control | 6.35E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC47A1 in breast cancer | [ 8 ] | |||
|
Location |
TSS1500 (cg12133118) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.08E+00 | Statistic Test | p-value:3.78E-02; Z-score:-7.59E-01 | ||
|
Methylation in Case |
5.23E-01 (Median) | Methylation in Control | 5.65E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC47A1 in breast cancer | [ 8 ] | |||
|
Location |
TSS200 (cg07829432) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:1.95E-05; Z-score:8.84E-01 | ||
|
Methylation in Case |
1.07E-01 (Median) | Methylation in Control | 8.72E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC47A1 in breast cancer | [ 8 ] | |||
|
Location |
TSS200 (cg13004927) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.35E+00 | Statistic Test | p-value:5.95E-04; Z-score:3.49E-01 | ||
|
Methylation in Case |
3.88E-02 (Median) | Methylation in Control | 2.88E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC47A1 in breast cancer | [ 8 ] | |||
|
Location |
TSS200 (cg15014549) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:6.71E-03; Z-score:4.96E-01 | ||
|
Methylation in Case |
7.76E-02 (Median) | Methylation in Control | 6.84E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC47A1 in breast cancer | [ 8 ] | |||
|
Location |
Body (cg08895056) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.28E+00 | Statistic Test | p-value:8.26E-13; Z-score:-2.23E+00 | ||
|
Methylation in Case |
5.37E-01 (Median) | Methylation in Control | 6.88E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC47A1 in breast cancer | [ 8 ] | |||
|
Location |
Body (cg20930201) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.45E+00 | Statistic Test | p-value:1.40E-10; Z-score:2.48E+00 | ||
|
Methylation in Case |
2.07E-01 (Median) | Methylation in Control | 1.43E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC47A1 in breast cancer | [ 8 ] | |||
|
Location |
Body (cg10718608) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.19E+00 | Statistic Test | p-value:7.64E-08; Z-score:-1.82E+00 | ||
|
Methylation in Case |
5.62E-01 (Median) | Methylation in Control | 6.67E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC47A1 in breast cancer | [ 8 ] | |||
|
Location |
Body (cg04308167) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.27E+00 | Statistic Test | p-value:1.27E-07; Z-score:-1.55E+00 | ||
|
Methylation in Case |
4.38E-01 (Median) | Methylation in Control | 5.57E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC47A1 in breast cancer | [ 8 ] | |||
|
Location |
Body (cg16887170) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.26E+00 | Statistic Test | p-value:4.94E-04; Z-score:-1.23E+00 | ||
|
Methylation in Case |
6.20E-01 (Median) | Methylation in Control | 7.83E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC47A1 in breast cancer | [ 8 ] | |||
|
Location |
Body (cg26959235) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.09E+00 | Statistic Test | p-value:5.02E-04; Z-score:4.52E-01 | ||
|
Methylation in Case |
1.29E-01 (Median) | Methylation in Control | 1.18E-01 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC47A1 in breast cancer | [ 8 ] | |||
|
Location |
Body (cg17332016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.06E+00 | Statistic Test | p-value:2.52E-03; Z-score:1.73E-01 | ||
|
Methylation in Case |
4.83E-02 (Median) | Methylation in Control | 4.55E-02 (Median) | ||
|
Studied Phenotype |
Breast cancer[ ICD-11:2C60-2C6Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Celiac disease |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC47A1 in celiac disease | [ 9 ] | |||
|
Location |
TSS1500 (cg21692194) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.61E+00 | Statistic Test | p-value:1.38E-03; Z-score:1.91E+00 | ||
|
Methylation in Case |
2.10E-01 (Median) | Methylation in Control | 1.31E-01 (Median) | ||
|
Studied Phenotype |
Celiac disease[ ICD-11:DA95] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC47A1 in celiac disease | [ 9 ] | |||
|
Location |
TSS1500 (cg25387636) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.84E+00 | Statistic Test | p-value:3.58E-02; Z-score:5.26E-01 | ||
|
Methylation in Case |
9.86E-02 (Median) | Methylation in Control | 5.37E-02 (Median) | ||
|
Studied Phenotype |
Celiac disease[ ICD-11:DA95] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC47A1 in celiac disease | [ 9 ] | |||
|
Location |
Body (cg16887170) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.22E+00 | Statistic Test | p-value:3.07E-02; Z-score:-1.14E+00 | ||
|
Methylation in Case |
7.01E-01 (Median) | Methylation in Control | 8.55E-01 (Median) | ||
|
Studied Phenotype |
Celiac disease[ ICD-11:DA95] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Colorectal cancer |
18 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC47A1 in colorectal cancer | [ 10 ] | |||
|
Location |
TSS1500 (cg15971010) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.20E+00 | Statistic Test | p-value:1.59E-12; Z-score:4.44E+00 | ||
|
Methylation in Case |
4.89E-01 (Median) | Methylation in Control | 2.22E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC47A1 in colorectal cancer | [ 10 ] | |||
|
Location |
TSS1500 (cg25387636) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.95E+00 | Statistic Test | p-value:2.49E-11; Z-score:6.62E+00 | ||
|
Methylation in Case |
3.66E-01 (Median) | Methylation in Control | 1.24E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC47A1 in colorectal cancer | [ 10 ] | |||
|
Location |
TSS1500 (cg21692194) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.91E+00 | Statistic Test | p-value:1.67E-09; Z-score:3.73E+00 | ||
|
Methylation in Case |
3.04E-01 (Median) | Methylation in Control | 1.59E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC47A1 in colorectal cancer | [ 10 ] | |||
|
Location |
TSS1500 (cg01530032) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:1.31E-08; Z-score:-2.16E+00 | ||
|
Methylation in Case |
7.03E-01 (Median) | Methylation in Control | 7.80E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC47A1 in colorectal cancer | [ 10 ] | |||
|
Location |
TSS200 (cg07829432) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.82E+00 | Statistic Test | p-value:2.59E-09; Z-score:3.90E+00 | ||
|
Methylation in Case |
3.13E-01 (Median) | Methylation in Control | 1.71E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC47A1 in colorectal cancer | [ 10 ] | |||
|
Location |
TSS200 (cg15014549) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.64E+00 | Statistic Test | p-value:1.01E-08; Z-score:3.34E+00 | ||
|
Methylation in Case |
1.91E-01 (Median) | Methylation in Control | 1.17E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC47A1 in colorectal cancer | [ 10 ] | |||
|
Location |
TSS200 (cg13004927) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.17E+00 | Statistic Test | p-value:3.99E-08; Z-score:1.58E+00 | ||
|
Methylation in Case |
6.96E-02 (Median) | Methylation in Control | 3.21E-02 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC47A1 in colorectal cancer | [ 10 ] | |||
|
Location |
Body (cg24151087) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.34E+00 | Statistic Test | p-value:1.12E-06; Z-score:1.88E+00 | ||
|
Methylation in Case |
4.17E-01 (Median) | Methylation in Control | 3.12E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC47A1 in colorectal cancer | [ 10 ] | |||
|
Location |
Body (cg26959235) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.29E+00 | Statistic Test | p-value:1.15E-06; Z-score:1.32E+00 | ||
|
Methylation in Case |
1.71E-01 (Median) | Methylation in Control | 1.32E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC47A1 in colorectal cancer | [ 10 ] | |||
|
Location |
Body (cg04308167) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.07E+00 | Statistic Test | p-value:2.25E-06; Z-score:-1.17E+00 | ||
|
Methylation in Case |
7.72E-01 (Median) | Methylation in Control | 8.29E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC47A1 in colorectal cancer | [ 10 ] | |||
|
Location |
Body (cg20930201) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.40E+00 | Statistic Test | p-value:3.29E-06; Z-score:1.80E+00 | ||
|
Methylation in Case |
3.26E-01 (Median) | Methylation in Control | 2.34E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon12 |
Methylation of SLC47A1 in colorectal cancer | [ 10 ] | |||
|
Location |
Body (cg17332016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:7.25E-05; Z-score:2.14E-01 | ||
|
Methylation in Case |
1.37E-01 (Median) | Methylation in Control | 1.32E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon13 |
Methylation of SLC47A1 in colorectal cancer | [ 10 ] | |||
|
Location |
Body (cg10718608) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:5.08E-04; Z-score:-7.26E-01 | ||
|
Methylation in Case |
8.24E-01 (Median) | Methylation in Control | 8.62E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon14 |
Methylation of SLC47A1 in colorectal cancer | [ 10 ] | |||
|
Location |
Body (cg11784214) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:8.98E-04; Z-score:-9.82E-01 | ||
|
Methylation in Case |
9.32E-01 (Median) | Methylation in Control | 9.41E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon15 |
Methylation of SLC47A1 in colorectal cancer | [ 10 ] | |||
|
Location |
Body (cg08895056) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.04E+00 | Statistic Test | p-value:9.10E-04; Z-score:-6.30E-01 | ||
|
Methylation in Case |
7.92E-01 (Median) | Methylation in Control | 8.20E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon16 |
Methylation of SLC47A1 in colorectal cancer | [ 10 ] | |||
|
Location |
Body (cg16887170) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.94E-02; Z-score:-4.04E-01 | ||
|
Methylation in Case |
8.71E-01 (Median) | Methylation in Control | 8.81E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon17 |
Methylation of SLC47A1 in colorectal cancer | [ 10 ] | |||
|
Location |
Body (cg12799818) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.01E+00 | Statistic Test | p-value:4.17E-02; Z-score:9.34E-02 | ||
|
Methylation in Case |
2.71E-01 (Median) | Methylation in Control | 2.67E-01 (Median) | ||
|
Studied Phenotype |
Colorectal cancer[ ICD-11:2B91] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
HIV infection |
11 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC47A1 in HIV infection | [ 11 ] | |||
|
Location |
TSS1500 (cg15971010) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.25E+00 | Statistic Test | p-value:4.88E-03; Z-score:6.76E-01 | ||
|
Methylation in Case |
1.70E-01 (Median) | Methylation in Control | 1.36E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC47A1 in HIV infection | [ 11 ] | |||
|
Location |
TSS1500 (cg25387636) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.32E+00 | Statistic Test | p-value:5.93E-03; Z-score:9.43E-01 | ||
|
Methylation in Case |
9.77E-02 (Median) | Methylation in Control | 7.37E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC47A1 in HIV infection | [ 11 ] | |||
|
Location |
TSS200 (cg07829432) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.34E+00 | Statistic Test | p-value:1.21E-04; Z-score:1.43E+00 | ||
|
Methylation in Case |
1.14E-01 (Median) | Methylation in Control | 8.46E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC47A1 in HIV infection | [ 11 ] | |||
|
Location |
TSS200 (cg13004927) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:2.65E+00 | Statistic Test | p-value:1.73E-03; Z-score:2.20E+00 | ||
|
Methylation in Case |
4.52E-02 (Median) | Methylation in Control | 1.71E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC47A1 in HIV infection | [ 11 ] | |||
|
Location |
Body (cg24151087) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.49E+00 | Statistic Test | p-value:2.12E-06; Z-score:2.25E+00 | ||
|
Methylation in Case |
1.91E-01 (Median) | Methylation in Control | 1.28E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC47A1 in HIV infection | [ 11 ] | |||
|
Location |
Body (cg16887170) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:1.11E-05; Z-score:-2.19E+00 | ||
|
Methylation in Case |
7.61E-01 (Median) | Methylation in Control | 8.36E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC47A1 in HIV infection | [ 11 ] | |||
|
Location |
Body (cg12799818) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.50E+00 | Statistic Test | p-value:2.09E-05; Z-score:1.85E+00 | ||
|
Methylation in Case |
8.68E-02 (Median) | Methylation in Control | 5.78E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC47A1 in HIV infection | [ 11 ] | |||
|
Location |
Body (cg20930201) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.34E+00 | Statistic Test | p-value:4.18E-05; Z-score:2.31E+00 | ||
|
Methylation in Case |
2.32E-01 (Median) | Methylation in Control | 1.74E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon9 |
Methylation of SLC47A1 in HIV infection | [ 11 ] | |||
|
Location |
Body (cg12894055) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.25E+00 | Statistic Test | p-value:4.43E-04; Z-score:8.90E-01 | ||
|
Methylation in Case |
5.22E-02 (Median) | Methylation in Control | 4.18E-02 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon10 |
Methylation of SLC47A1 in HIV infection | [ 11 ] | |||
|
Location |
Body (cg26959235) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.19E+00 | Statistic Test | p-value:1.57E-02; Z-score:7.72E-01 | ||
|
Methylation in Case |
1.45E-01 (Median) | Methylation in Control | 1.22E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon11 |
Methylation of SLC47A1 in HIV infection | [ 11 ] | |||
|
Location |
Body (cg10718608) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.18E+00 | Statistic Test | p-value:3.00E-02; Z-score:1.30E+00 | ||
|
Methylation in Case |
3.15E-01 (Median) | Methylation in Control | 2.67E-01 (Median) | ||
|
Studied Phenotype |
HIV infection[ ICD-11:1C62.Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Prostate cancer |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC47A1 in prostate cancer | [ 12 ] | |||
|
Location |
TSS1500 (cg20533553) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:2.38E-02; Z-score:2.62E+00 | ||
|
Methylation in Case |
7.79E-01 (Median) | Methylation in Control | 6.38E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC47A1 in prostate cancer | [ 12 ] | |||
|
Location |
Body (cg08874645) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.17E+00 | Statistic Test | p-value:1.34E-02; Z-score:1.88E+00 | ||
|
Methylation in Case |
8.36E-01 (Median) | Methylation in Control | 7.15E-01 (Median) | ||
|
Studied Phenotype |
Prostate cancer[ ICD-11:2C82] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Renal cell carcinoma |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC47A1 in clear cell renal cell carcinoma | [ 13 ] | |||
|
Location |
TSS200 (cg15014549) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.13E+00 | Statistic Test | p-value:1.50E-02; Z-score:5.02E-01 | ||
|
Methylation in Case |
3.47E-02 (Median) | Methylation in Control | 3.06E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC47A1 in clear cell renal cell carcinoma | [ 13 ] | |||
|
Location |
TSS200 (cg07829432) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.14E+00 | Statistic Test | p-value:3.16E-02; Z-score:2.64E-01 | ||
|
Methylation in Case |
3.80E-02 (Median) | Methylation in Control | 3.34E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC47A1 in clear cell renal cell carcinoma | [ 13 ] | |||
|
Location |
Body (cg12894055) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-2.41E+00 | Statistic Test | p-value:6.34E-08; Z-score:-2.52E+00 | ||
|
Methylation in Case |
5.62E-02 (Median) | Methylation in Control | 1.35E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC47A1 in clear cell renal cell carcinoma | [ 13 ] | |||
|
Location |
Body (cg10718608) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.32E+00 | Statistic Test | p-value:3.14E-07; Z-score:-3.83E+00 | ||
|
Methylation in Case |
5.26E-01 (Median) | Methylation in Control | 6.94E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC47A1 in clear cell renal cell carcinoma | [ 13 ] | |||
|
Location |
Body (cg08895056) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.32E+00 | Statistic Test | p-value:1.59E-06; Z-score:-3.32E+00 | ||
|
Methylation in Case |
5.11E-01 (Median) | Methylation in Control | 6.73E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC47A1 in clear cell renal cell carcinoma | [ 13 ] | |||
|
Location |
Body (cg20930201) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.36E+00 | Statistic Test | p-value:4.54E-03; Z-score:9.20E-01 | ||
|
Methylation in Case |
1.07E-01 (Median) | Methylation in Control | 7.89E-02 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC47A1 in clear cell renal cell carcinoma | [ 13 ] | |||
|
Location |
3'UTR (cg12550399) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.12E+00 | Statistic Test | p-value:2.75E-03; Z-score:2.21E+00 | ||
|
Methylation in Case |
8.54E-01 (Median) | Methylation in Control | 7.64E-01 (Median) | ||
|
Studied Phenotype |
Renal cell carcinoma[ ICD-11:2C90] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Lung adenocarcinoma |
8 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC47A1 in lung adenocarcinoma | [ 14 ] | |||
|
Location |
TSS200 (cg07829432) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.46E+00 | Statistic Test | p-value:3.51E-03; Z-score:3.46E+00 | ||
|
Methylation in Case |
1.84E-01 (Median) | Methylation in Control | 1.26E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC47A1 in lung adenocarcinoma | [ 14 ] | |||
|
Location |
TSS200 (cg15014549) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.20E+00 | Statistic Test | p-value:7.76E-03; Z-score:2.65E+00 | ||
|
Methylation in Case |
1.25E-01 (Median) | Methylation in Control | 1.04E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC47A1 in lung adenocarcinoma | [ 14 ] | |||
|
Location |
TSS200 (cg13004927) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.62E+00 | Statistic Test | p-value:4.44E-02; Z-score:1.78E+00 | ||
|
Methylation in Case |
7.07E-02 (Median) | Methylation in Control | 4.38E-02 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC47A1 in lung adenocarcinoma | [ 14 ] | |||
|
Location |
Body (cg10718608) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.29E+00 | Statistic Test | p-value:5.10E-05; Z-score:-3.35E+00 | ||
|
Methylation in Case |
4.90E-01 (Median) | Methylation in Control | 6.34E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC47A1 in lung adenocarcinoma | [ 14 ] | |||
|
Location |
Body (cg08895056) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.23E+00 | Statistic Test | p-value:1.04E-04; Z-score:-2.47E+00 | ||
|
Methylation in Case |
5.25E-01 (Median) | Methylation in Control | 6.48E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC47A1 in lung adenocarcinoma | [ 14 ] | |||
|
Location |
Body (cg12894055) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.49E+00 | Statistic Test | p-value:5.84E-03; Z-score:-1.48E+00 | ||
|
Methylation in Case |
1.49E-01 (Median) | Methylation in Control | 2.22E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC47A1 in lung adenocarcinoma | [ 14 ] | |||
|
Location |
Body (cg16887170) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:7.22E-03; Z-score:-1.53E+00 | ||
|
Methylation in Case |
7.68E-01 (Median) | Methylation in Control | 8.13E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC47A1 in lung adenocarcinoma | [ 14 ] | |||
|
Location |
Body (cg20930201) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.16E+00 | Statistic Test | p-value:2.61E-02; Z-score:1.29E+00 | ||
|
Methylation in Case |
2.47E-01 (Median) | Methylation in Control | 2.13E-01 (Median) | ||
|
Studied Phenotype |
Lung adenocarcinoma[ ICD-11:2C25.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Panic disorder |
3 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC47A1 in panic disorder | [ 15 ] | |||
|
Location |
TSS200 (cg15014549) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:9.75E-01 | Statistic Test | p-value:6.59E-03; Z-score:3.02E-01 | ||
|
Methylation in Case |
-4.09E+00 (Median) | Methylation in Control | -4.19E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC47A1 in panic disorder | [ 15 ] | |||
|
Location |
Body (cg17332016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-9.80E-01 | Statistic Test | p-value:2.59E-02; Z-score:-3.18E-01 | ||
|
Methylation in Case |
-5.20E+00 (Median) | Methylation in Control | -5.10E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC47A1 in panic disorder | [ 15 ] | |||
|
Location |
Body (cg10718608) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:9.46E-01 | Statistic Test | p-value:4.36E-02; Z-score:3.36E-01 | ||
|
Methylation in Case |
-1.97E+00 (Median) | Methylation in Control | -2.09E+00 (Median) | ||
|
Studied Phenotype |
Panic disorder[ ICD-11:6B01] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Atypical teratoid rhabdoid tumor |
7 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC47A1 in atypical teratoid rhabdoid tumor | [ 16 ] | |||
|
Location |
Body (cg04308167) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.29E+00 | Statistic Test | p-value:6.87E-04; Z-score:-9.00E-01 | ||
|
Methylation in Case |
3.86E-01 (Median) | Methylation in Control | 4.96E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC47A1 in atypical teratoid rhabdoid tumor | [ 16 ] | |||
|
Location |
Body (cg08895056) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.04E+00 | Statistic Test | p-value:8.62E-03; Z-score:4.86E-01 | ||
|
Methylation in Case |
9.34E-01 (Median) | Methylation in Control | 9.02E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC47A1 in atypical teratoid rhabdoid tumor | [ 16 ] | |||
|
Location |
Body (cg10718608) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.05E+00 | Statistic Test | p-value:1.75E-02; Z-score:-3.83E-01 | ||
|
Methylation in Case |
7.59E-01 (Median) | Methylation in Control | 7.96E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC47A1 in atypical teratoid rhabdoid tumor | [ 16 ] | |||
|
Location |
Body (cg11784214) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.51E+00 | Statistic Test | p-value:2.49E-02; Z-score:-6.29E-01 | ||
|
Methylation in Case |
1.74E-01 (Median) | Methylation in Control | 2.62E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC47A1 in atypical teratoid rhabdoid tumor | [ 16 ] | |||
|
Location |
Body (cg12799818) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.22E+00 | Statistic Test | p-value:2.97E-02; Z-score:8.34E-01 | ||
|
Methylation in Case |
6.40E-01 (Median) | Methylation in Control | 5.27E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC47A1 in atypical teratoid rhabdoid tumor | [ 16 ] | |||
|
Location |
Body (cg12894055) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:1.05E+00 | Statistic Test | p-value:3.11E-02; Z-score:3.82E-01 | ||
|
Methylation in Case |
8.83E-01 (Median) | Methylation in Control | 8.39E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC47A1 in atypical teratoid rhabdoid tumor | [ 16 ] | |||
|
Location |
3'UTR (cg12550399) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.60E+00 | Statistic Test | p-value:1.55E-11; Z-score:-1.75E+00 | ||
|
Methylation in Case |
2.62E-01 (Median) | Methylation in Control | 4.19E-01 (Median) | ||
|
Studied Phenotype |
Atypical teratoid rhabdoid tumor[ ICD-11:2A00.1Y] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Depression |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC47A1 in depression | [ 17 ] | |||
|
Location |
Body (cg16887170) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.02E+00 | Statistic Test | p-value:1.07E-03; Z-score:-6.47E-01 | ||
|
Methylation in Case |
7.63E-01 (Median) | Methylation in Control | 7.80E-01 (Median) | ||
|
Studied Phenotype |
Depression[ ICD-11:6A8Z] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Papillary thyroid cancer |
8 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC47A1 in papillary thyroid cancer | [ 18 ] | |||
|
Location |
Body (cg12894055) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.65E+00 | Statistic Test | p-value:2.07E-17; Z-score:-1.68E+00 | ||
|
Methylation in Case |
9.77E-02 (Median) | Methylation in Control | 1.61E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC47A1 in papillary thyroid cancer | [ 18 ] | |||
|
Location |
Body (cg26959235) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.26E+00 | Statistic Test | p-value:5.83E-16; Z-score:-1.96E+00 | ||
|
Methylation in Case |
1.52E-01 (Median) | Methylation in Control | 1.91E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC47A1 in papillary thyroid cancer | [ 18 ] | |||
|
Location |
Body (cg24151087) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.32E+00 | Statistic Test | p-value:1.46E-13; Z-score:-2.20E+00 | ||
|
Methylation in Case |
4.10E-01 (Median) | Methylation in Control | 5.41E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC47A1 in papillary thyroid cancer | [ 18 ] | |||
|
Location |
Body (cg08895056) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.32E+00 | Statistic Test | p-value:3.58E-12; Z-score:-1.97E+00 | ||
|
Methylation in Case |
3.56E-01 (Median) | Methylation in Control | 4.70E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon5 |
Methylation of SLC47A1 in papillary thyroid cancer | [ 18 ] | |||
|
Location |
Body (cg10718608) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.25E+00 | Statistic Test | p-value:1.10E-10; Z-score:-1.78E+00 | ||
|
Methylation in Case |
3.68E-01 (Median) | Methylation in Control | 4.60E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon6 |
Methylation of SLC47A1 in papillary thyroid cancer | [ 18 ] | |||
|
Location |
Body (cg12799818) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.34E+00 | Statistic Test | p-value:3.35E-07; Z-score:-1.56E+00 | ||
|
Methylation in Case |
2.92E-01 (Median) | Methylation in Control | 3.90E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon7 |
Methylation of SLC47A1 in papillary thyroid cancer | [ 18 ] | |||
|
Location |
Body (cg17332016) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.10E+00 | Statistic Test | p-value:1.43E-03; Z-score:-4.84E-01 | ||
|
Methylation in Case |
4.25E-02 (Median) | Methylation in Control | 4.68E-02 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon8 |
Methylation of SLC47A1 in papillary thyroid cancer | [ 18 ] | |||
|
Location |
Body (cg04308167) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.03E+00 | Statistic Test | p-value:1.31E-02; Z-score:-4.65E-01 | ||
|
Methylation in Case |
7.60E-01 (Median) | Methylation in Control | 7.85E-01 (Median) | ||
|
Studied Phenotype |
Papillary thyroid cancer[ ICD-11:2D10.1] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Systemic lupus erythematosus |
4 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Methylation of SLC47A1 in systemic lupus erythematosus | [ 19 ] | |||
|
Location |
Body (cg04308167) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.11E+00 | Statistic Test | p-value:6.32E-03; Z-score:-3.99E-01 | ||
|
Methylation in Case |
4.89E-01 (Median) | Methylation in Control | 5.44E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon2 |
Methylation of SLC47A1 in systemic lupus erythematosus | [ 19 ] | |||
|
Location |
Body (cg11784214) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:1.40E-02; Z-score:-1.44E-01 | ||
|
Methylation in Case |
8.30E-01 (Median) | Methylation in Control | 8.35E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon3 |
Methylation of SLC47A1 in systemic lupus erythematosus | [ 19 ] | |||
|
Location |
Body (cg12894055) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.06E+00 | Statistic Test | p-value:1.79E-02; Z-score:-1.01E-01 | ||
|
Methylation in Case |
7.10E-02 (Median) | Methylation in Control | 7.53E-02 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Epigenetic Phenomenon4 |
Methylation of SLC47A1 in systemic lupus erythematosus | [ 19 ] | |||
|
Location |
Body (cg16887170) | ||||
|
Epigenetic Type |
Methylation | Experiment Method | Infinium HumanMethylation450 BeadChip | ||
|
Methylation Fold Change |
Fold Change:-1.01E+00 | Statistic Test | p-value:3.38E-02; Z-score:-8.87E-02 | ||
|
Methylation in Case |
7.87E-01 (Median) | Methylation in Control | 7.94E-01 (Median) | ||
|
Studied Phenotype |
Systemic lupus erythematosus[ ICD-11:4A40.0] | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Gastric cancer |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate/moderate hypermethylation of SLC47A1 in gastric cancer than that in healthy individual/adjacent tissue | ||||
Studied Phenotype |
Gastric cancer [ICD-11:2B72] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:4.98E-15; Fold-change:0.209868716; Z-score:1.624788814 | ||||
The Methylation Level of Disease Section Compare with the Adjacent Tissue |
p-value:8.21E-66; Fold-change:0.284929054; Z-score:170.8903475 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue adjacent to the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
|
Prostate cancer metastasis |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
Moderate hypermethylation of SLC47A1 in prostate cancer metastasis than that in healthy individual | ||||
Studied Phenotype |
Prostate cancer metastasis [ICD-11:2.00E+06] | ||||
The Methylation Level of Disease Section Compare with the Healthy Individual |
p-value:0.037135313; Fold-change:0.242405384; Z-score:9.410363493 | ||||
| DT methylation level in the diseased tissue of patients
DT methylation level in the normal tissue of healthy individuals
|
|||||
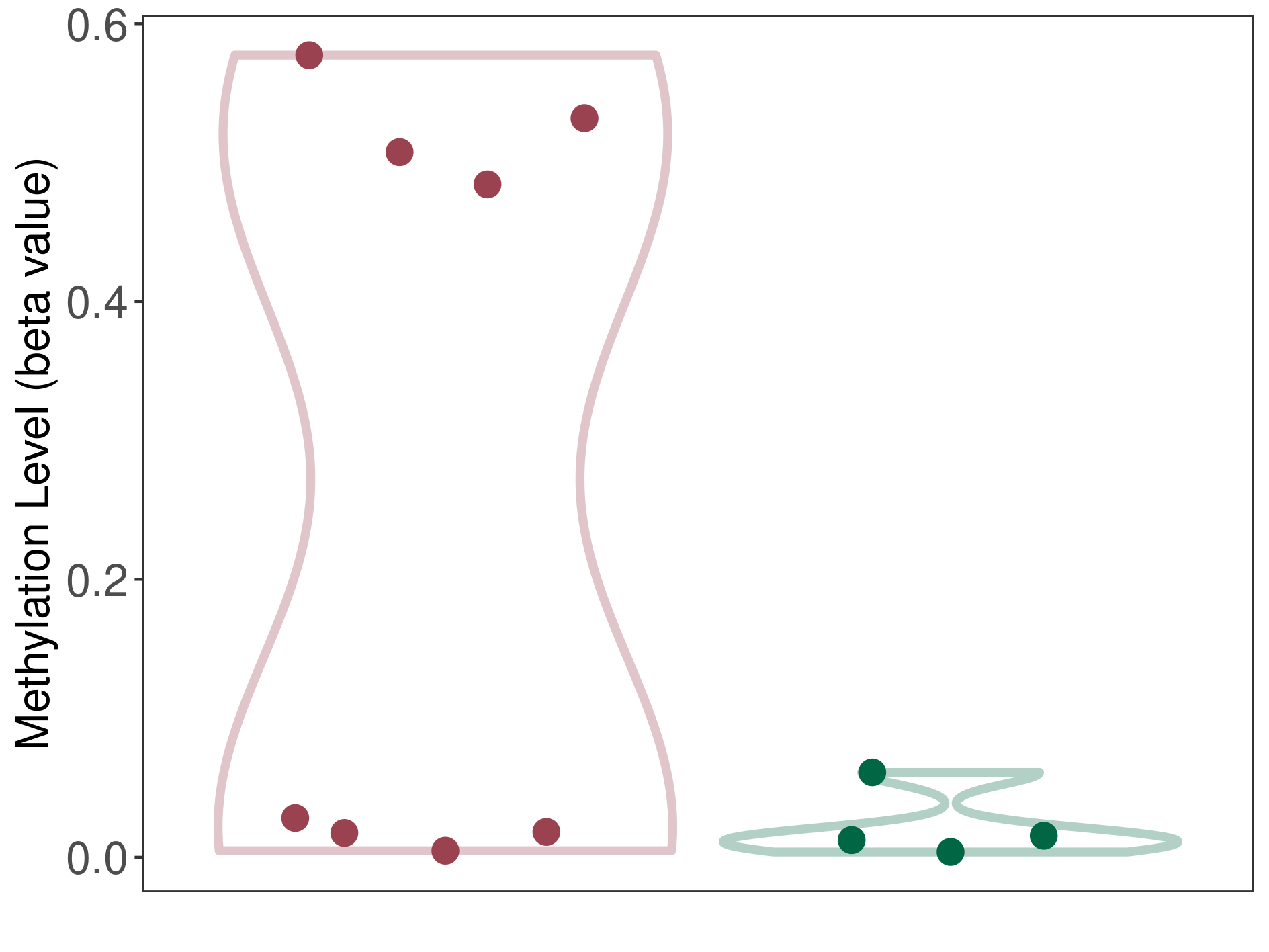
|
 Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
Please Click the above Thumbnail to View/Download
the Methylation Barchart for All Samples
| ||||
|
microRNA |
|||||
|
Unclear Phenotype |
52 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
miR-1275 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1275 | miRNA Mature ID | miR-1275 | ||
|
miRNA Sequence |
GUGGGGGAGAGGCUGUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon2 |
miR-1307 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1307 | miRNA Mature ID | miR-1307-3p | ||
|
miRNA Sequence |
ACUCGGCGUGGCGUCGGUCGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon3 |
miR-1343 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1343 | miRNA Mature ID | miR-1343-5p | ||
|
miRNA Sequence |
UGGGGAGCGGCCCCCGGGUGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon4 |
miR-149 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-149 | miRNA Mature ID | miR-149-3p | ||
|
miRNA Sequence |
AGGGAGGGACGGGGGCUGUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon5 |
miR-181d directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-181d | miRNA Mature ID | miR-181d-3p | ||
|
miRNA Sequence |
CCACCGGGGGAUGAAUGUCAC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon6 |
miR-1908 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-1908 | miRNA Mature ID | miR-1908-5p | ||
|
miRNA Sequence |
CGGCGGGGACGGCGAUUGGUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon7 |
miR-212 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-212 | miRNA Mature ID | miR-212-5p | ||
|
miRNA Sequence |
ACCUUGGCUCUAGACUGCUUACU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon8 |
miR-3144 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3144 | miRNA Mature ID | miR-3144-5p | ||
|
miRNA Sequence |
AGGGGACCAAAGAGAUAUAUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon9 |
miR-3189 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3189 | miRNA Mature ID | miR-3189-3p | ||
|
miRNA Sequence |
CCCUUGGGUCUGAUGGGGUAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon10 |
miR-326 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-326 | miRNA Mature ID | miR-326 | ||
|
miRNA Sequence |
CCUCUGGGCCCUUCCUCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon11 |
miR-330 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-330 | miRNA Mature ID | miR-330-5p | ||
|
miRNA Sequence |
UCUCUGGGCCUGUGUCUUAGGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon12 |
miR-3616 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3616 | miRNA Mature ID | miR-3616-3p | ||
|
miRNA Sequence |
CGAGGGCAUUUCAUGAUGCAGGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon13 |
miR-3960 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-3960 | miRNA Mature ID | miR-3960 | ||
|
miRNA Sequence |
GGCGGCGGCGGAGGCGGGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon14 |
miR-4300 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4300 | miRNA Mature ID | miR-4300 | ||
|
miRNA Sequence |
UGGGAGCUGGACUACUUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon15 |
miR-4467 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4467 | miRNA Mature ID | miR-4467 | ||
|
miRNA Sequence |
UGGCGGCGGUAGUUAUGGGCUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon16 |
miR-4525 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4525 | miRNA Mature ID | miR-4525 | ||
|
miRNA Sequence |
GGGGGGAUGUGCAUGCUGGUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon17 |
miR-4655 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4655 | miRNA Mature ID | miR-4655-5p | ||
|
miRNA Sequence |
CACCGGGGAUGGCAGAGGGUCG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon18 |
miR-4665 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4665 | miRNA Mature ID | miR-4665-5p | ||
|
miRNA Sequence |
CUGGGGGACGCGUGAGCGCGAGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon19 |
miR-4667 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4667 | miRNA Mature ID | miR-4667-5p | ||
|
miRNA Sequence |
ACUGGGGAGCAGAAGGAGAACC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon20 |
miR-4680 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4680 | miRNA Mature ID | miR-4680-5p | ||
|
miRNA Sequence |
AGAACUCUUGCAGUCUUAGAUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon21 |
miR-4700 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4700 | miRNA Mature ID | miR-4700-5p | ||
|
miRNA Sequence |
UCUGGGGAUGAGGACAGUGUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon22 |
miR-4706 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4706 | miRNA Mature ID | miR-4706 | ||
|
miRNA Sequence |
AGCGGGGAGGAAGUGGGCGCUGCUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon23 |
miR-4723 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4723 | miRNA Mature ID | miR-4723-5p | ||
|
miRNA Sequence |
UGGGGGAGCCAUGAGAUAAGAGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon24 |
miR-4728 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4728 | miRNA Mature ID | miR-4728-5p | ||
|
miRNA Sequence |
UGGGAGGGGAGAGGCAGCAAGCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon25 |
miR-4730 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4730 | miRNA Mature ID | miR-4730 | ||
|
miRNA Sequence |
CUGGCGGAGCCCAUUCCAUGCCA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon26 |
miR-4749 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-4749 | miRNA Mature ID | miR-4749-5p | ||
|
miRNA Sequence |
UGCGGGGACAGGCCAGGGCAUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon27 |
miR-5008 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-5008 | miRNA Mature ID | miR-5008-5p | ||
|
miRNA Sequence |
UGAGGCCCUUGGGGCACAGUGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon28 |
miR-5010 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-5010 | miRNA Mature ID | miR-5010-5p | ||
|
miRNA Sequence |
AGGGGGAUGGCAGAGCAAAAUU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon29 |
miR-514a directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-514a | miRNA Mature ID | miR-514a-5p | ||
|
miRNA Sequence |
UACUCUGGAGAGUGACAAUCAUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon30 |
miR-5591 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-5591 | miRNA Mature ID | miR-5591-5p | ||
|
miRNA Sequence |
UGGGAGCUAAGCUAUGGGUAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon31 |
miR-5698 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-5698 | miRNA Mature ID | miR-5698 | ||
|
miRNA Sequence |
UGGGGGAGUGCAGUGAUUGUGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon32 |
miR-625 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-625 | miRNA Mature ID | miR-625-5p | ||
|
miRNA Sequence |
AGGGGGAAAGUUCUAUAGUCC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon33 |
miR-637 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-637 | miRNA Mature ID | miR-637 | ||
|
miRNA Sequence |
ACUGGGGGCUUUCGGGCUCUGCGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon34 |
miR-663a directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-663a | miRNA Mature ID | miR-663a | ||
|
miRNA Sequence |
AGGCGGGGCGCCGCGGGACCGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon35 |
miR-6726 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6726 | miRNA Mature ID | miR-6726-5p | ||
|
miRNA Sequence |
CGGGAGCUGGGGUCUGCAGGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon36 |
miR-6770 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6770 | miRNA Mature ID | miR-6770-3p | ||
|
miRNA Sequence |
CUGGCGGCUGUGUCUUCACAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon37 |
miR-6777 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6777 | miRNA Mature ID | miR-6777-5p | ||
|
miRNA Sequence |
ACGGGGAGUCAGGCAGUGGUGGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon38 |
miR-6784 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6784 | miRNA Mature ID | miR-6784-5p | ||
|
miRNA Sequence |
GCCGGGGCUUUGGGUGAGGG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon39 |
miR-6785 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6785 | miRNA Mature ID | miR-6785-5p | ||
|
miRNA Sequence |
UGGGAGGGCGUGGAUGAUGGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon40 |
miR-6787 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6787 | miRNA Mature ID | miR-6787-5p | ||
|
miRNA Sequence |
UGGCGGGGGUAGAGCUGGCUGC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon41 |
miR-6825 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6825 | miRNA Mature ID | miR-6825-5p | ||
|
miRNA Sequence |
UGGGGAGGUGUGGAGUCAGCAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon42 |
miR-6851 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6851 | miRNA Mature ID | miR-6851-3p | ||
|
miRNA Sequence |
UGGCCCUUUGUACCCCUCCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon43 |
miR-6870 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6870 | miRNA Mature ID | miR-6870-5p | ||
|
miRNA Sequence |
UGGGGGAGAUGGGGGUUGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon44 |
miR-6883 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6883 | miRNA Mature ID | miR-6883-5p | ||
|
miRNA Sequence |
AGGGAGGGUGUGGUAUGGAUGU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon45 |
miR-6889 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-6889 | miRNA Mature ID | miR-6889-5p | ||
|
miRNA Sequence |
UCGGGGAGUCUGGGGUCCGGAAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon46 |
miR-7111 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-7111 | miRNA Mature ID | miR-7111-5p | ||
|
miRNA Sequence |
UGGGGGAGGAAGGACAGGCCAU | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon47 |
miR-7155 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-7155 | miRNA Mature ID | miR-7155-5p | ||
|
miRNA Sequence |
UCUGGGGUCUUGGGCCAUC | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon48 |
miR-7160 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-7160 | miRNA Mature ID | miR-7160-3p | ||
|
miRNA Sequence |
CAGGGCCCUGGCUUUAGCAGA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon49 |
miR-8072 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-8072 | miRNA Mature ID | miR-8072 | ||
|
miRNA Sequence |
GGCGGCGGGGAGGUAGGCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon50 |
miR-8089 directly targets SLC47A1 | [ 20 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-8089 | miRNA Mature ID | miR-8089 | ||
|
miRNA Sequence |
CCUGGGGACAGGGGAUUGGGGCAG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Human embryonic kidney 293 cells (HEK293) | ||||
|
Epigenetic Phenomenon51 |
miR-920 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-920 | miRNA Mature ID | miR-920 | ||
|
miRNA Sequence |
GGGGAGCUGUGGAAGCAGUA | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Epigenetic Phenomenon52 |
miR-939 directly targets SLC47A1 | [ 21 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | PAR-CLIP | ||
|
miRNA Stemloop ID |
miR-939 | miRNA Mature ID | miR-939-5p | ||
|
miRNA Sequence |
UGGGGAGCUGAGGCUCUGGGGGUG | ||||
|
miRNA Target Type |
Direct | ||||
|
Experimental Material |
Multiple cell lines of human | ||||
|
Human liver tissue |
1 Epigenetic Phenomena Related to This Phenotype | Click to Show/Hide the Full List | |||
|
Epigenetic Phenomenon1 |
miR-95 regulates SLC47A1 expression | [ 22 ] | |||
|
Epigenetic Type |
microRNA | Experiment Method | Taqman OpenArray Human miRNA Panel | ||
|
miRNA Stemloop ID |
miR-95 | miRNA Mature ID | miR-95-5p | ||
|
miRNA Sequence |
UCAAUAAAUGUCUGUUGAAUU | ||||
|
miRNA Target Type |
Undirect | ||||
|
Studied Phenotype |
Human liver tissue | ||||
|
Experimental Material |
Patient tissue samples | ||||
|
Additional Notes |
The effects of rifampin on four uptake drug transporters SLC47A1 were negatively correlated with the rifampin effects on miR-95 expression(r=-0.79, p=0.0048). | ||||
If you find any error in data or bug in web service, please kindly report it to Dr. Li and Dr. Fu.
