Description of data format
1. Required Data Formats in the Input Files
The file required at the beginning of NOREVA analysis should provide a sample-by-feature matrix in a csv format. The required input file in correct format could be readily generated based on results of several popular tools such as XCMS online.
Input File for Analyzing Metabolomics Data with QC Samples
In this situation, the sample ID, batch ID, class of samples and injection order are sequentially provided in the first four columns of input file. Names of these columns must be kept as “sample”, “batch”, “class” and “order” without any changes during the entire analysis. The sample ID is uniquely assigned according to users’ preference; the batch ID refers to different analytical blocks or batches, and is labeled with ordinal number, e.g., 1,2,3,…; the class of samples indicates two sample groups and QC samples (the name of sample groups is different, and QC samples are all labeled as “NA”); the injection order is strictly follow the sequence of experiment.

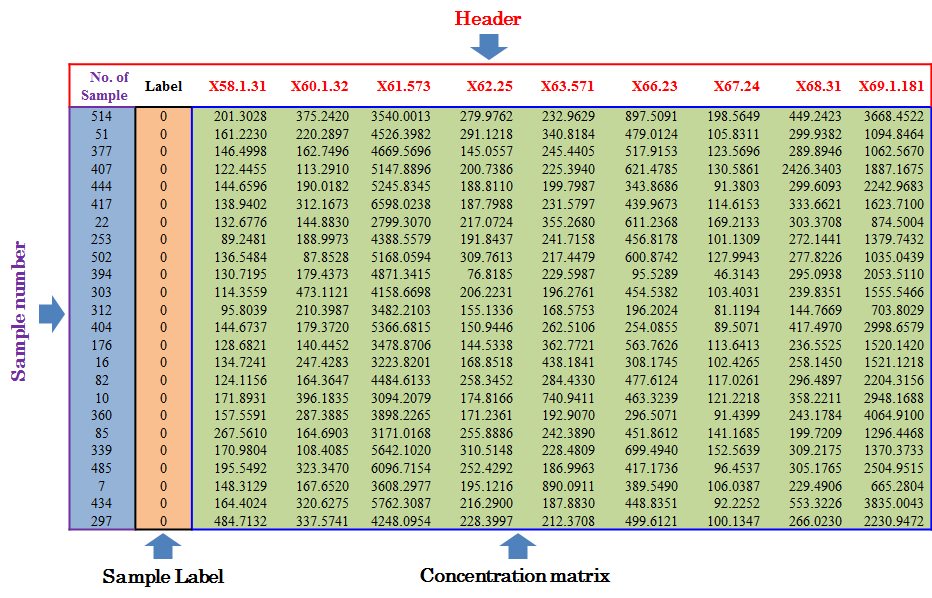
Input File for Analyzing Dataset without QC Samples or Experiments Ignoring QC Preparation
Under this circumstance, only sample ID and class of samples are required in the first 2 columns of the input file, and are kept as “SampleName” and “Label”. In the column of class of samples, “NA” is not labeled to any sample due to the absence of QC samples. In the following columns of both types of input file, metabolites’ raw intensities across all samples without logarithmic scaling are further provided. Unique IDs of each metabolite are listed in the first row of the csv file.
Input File Containing Reference Metabolites as Golden Standards
To evaluate methods based on the last criterion, additional file providing information of the reference metabolites (e.g., spike-in compounds) is needed for further analysis. In this file, sample ID and class of samples are required in the first 2 columns. Their names are provided as “sample” and “class”. The sample ID is also uniquely assigned according to users’ preference, and the class of samples indicates two sample groups of different name. Example file can also be downloaded from NOREVA.
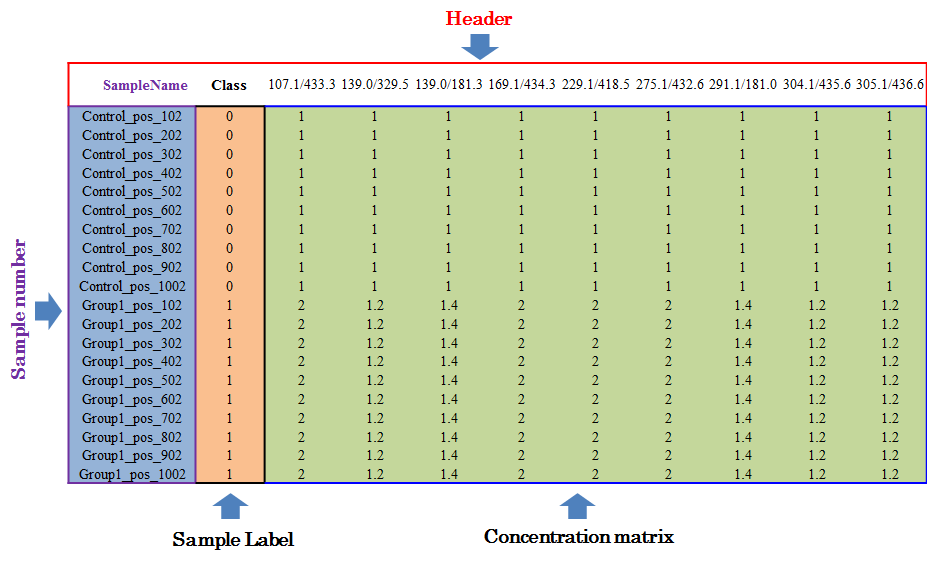
Input File for Analyzing Dataset including the internal standards or QC metabolites
Under this circumstance, only sample name and class of samples are required in the first 2 columns of the input file, and are kept as “SampleName” and “Label”. In the column of class of samples, “NA” is not labeled to any sample due to the absence of QC samples. In the following columns of both types of input file, metabolites’ raw intensities across all samples without logarithmic scaling are further provided. Unique IDs of each metabolite are listed in the first row of the csv file.
